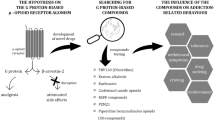
Abstract
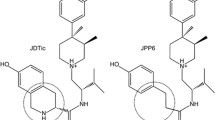
Opioids are potent painkillers, however, their therapeutic use requires close medical monitoring to diminish the risk of severe adverse effects. The G-protein biased agonists of the μ-opioid receptor (MOR) have shown safer therapeutic profiles than non-biased ligands. In this work, we performed extensive all-atom molecular dynamics simulations of two markedly biased ligands and a balanced reference molecule. From those simulations, we identified a protein–ligand interaction fingerprint that characterizes biased ligands. Then, we built and virtually screened a database containing 68,740 ligands with proven or potential GPCR agonistic activity. Exemplary molecules that fulfill the interacting pattern for biased agonism are showcased, illustrating the usefulness of this work for the search of biased MOR ligands and how this contributes to the understanding of MOR biased signaling.





Similar content being viewed by others
Data availability
Our manuscript has data included as electronic supplementary information.
Code availability
Not applicable.
References
Santos R et al (2016) A comprehensive map of molecular drug targets. Nat Rev Drug Discov 16(1):19–34. https://doi.org/10.1038/nrd.2016.230
Madariaga-Mazón A, Marmolejo-Valencia AF, Li Y, Toll L, Houghten RA, Martinez-Mayorga K (2017) Mu-Opioid receptor biased ligands: A safer and painless discovery of analgesics? Drug Discov Today 22(11):1719–1729. https://doi.org/10.1016/j.drudis.2017.07.002
Domazet I et al (2015) Characterization of angiotensin II molecular determinants involved in AT1 receptor functional selectivity. Mol Pharmacol 87(6):982–995. https://doi.org/10.1124/mol.114.097337
McCorvy JD et al (2018) Structural determinants of 5-HT2B receptor activation and biased agonism. Nat Struct Mol Biol 25(9):787–796. https://doi.org/10.1038/s41594-018-0116-7
Zhang H et al (2015) Structural basis for ligand recognition and functional selectivity at angiotensin receptor. J Biol Chem 290(49):29127–29139. https://doi.org/10.1074/jbc.M115.689000
Hothersall JD et al (2017) Residues W320 and Y328 within the binding site of the μ-opioid receptor influence opiate ligand bias. Neuropharmacology 118:46–58. https://doi.org/10.1016/j.neuropharm.2017.03.007
Cheng JX, Cheng T, Li WH, Liu GX, Zhu WL, Tang Y (2018) Computational insights into the G-protein-biased activation and inactivation mechanisms of the μ opioid receptor. Acta Pharmacol Sin 39(1):154–164. https://doi.org/10.1038/aps.2017.158
Huang W et al (2015) Structural insights into µ-opioid receptor activation. Nature. https://doi.org/10.1038/nature14886
Bermudez M et al (2017) Ligand-specific restriction of extracellular conformational dynamics constrains signaling of the M2 muscarinic receptor. ACS Chem Biol 12(7):1743–1748. https://doi.org/10.1021/acschembio.7b00275
Ring AM et al (2013) Adrenaline-activated structure of β 2-adrenoceptor stabilized by an engineered nanobody. Nature 502(7472):575–579. https://doi.org/10.1038/nature12572
Capper MJ, Wacker D (2018) Structural Biology: A complex story of receptor signalling. Nature 558(7711):529–530. https://doi.org/10.1038/d41586-018-05503-4
Devree BT et al (2016) Allosteric coupling from G protein to the agonist-binding pocket in GPCRs. Nature 535(7610):182–186. https://doi.org/10.1038/nature18324
Manglik A et al (2016) Structure-based discovery of opioid analgesics with reduced side effects. Nature 537(7619):185–190. https://doi.org/10.1038/nature19112
Bermudez M, Bock A (2019) Does divergent binding pocket closure drive ligand bias for class A GPCRs? Trends Pharmacol Sci 40(4):236–239. https://doi.org/10.1016/j.tips.2019.02.005
Liu W et al (2012) Structural basis for allosteric regulation of GPCRS by sodium ions. Science 337(6091):232–236. https://doi.org/10.1126/science.1219218
Yabaluri N, Medzihradsky F (2002) Regulation of μ-opioid receptor in neural cells by extracellular sodium. J Neurochem 68(3):1053–1061. https://doi.org/10.1046/j.1471-4159.1997.68031053.x
Pert CB, Pasternak G, Snyder SH (1973) Opiate agonists and antagonists discriminated by receptor binding in brain. Science 182(4119):1359–1361. https://doi.org/10.1126/science.182.4119.1359
Xu W et al (1999) Functional role of the spatial proximity of Asp114(2.50) in TMH 2 and Asn332(7.49) in TMH 7 of the μ opioid receptor. FEBS Lett 447(2–3):318–324. https://doi.org/10.1016/S0014-5793(99)00316-6
Fenalti G et al (2014) Molecular control of δ-opioid receptor signalling. Nature 506(7487):191–196. https://doi.org/10.1038/nature12944
Miller-Gallacher JL et al (2014) The 2.1 Å resolution structure of cyanopindolol-bound β1-adrenoceptor identifies an intramembrane Na+ ion that stabilises the ligand-free receptor. PLoS ONE. https://doi.org/10.1371/journal.pone.0092727
Hu X, Wang Y, Hunkele A, Provasi D, Pasternak GW, Filizola M (2019) Kinetic and thermodynamic insights into sodium ion translocation through the μ-opioid receptor from molecular dynamics and machine learning analysis. PLOS Comput Biol. https://doi.org/10.1371/journal.pcbi.1006689
Marmolejo-Valencia AF, Martínez-Mayorga K (2017) Allosteric modulation model of the mu opioid receptor by herkinorin, a potent not alkaloidal agonist. J Comput Aided Mol Des 31(5):467–482. https://doi.org/10.1007/s10822-017-0016-7
DeWire SM et al (2013) A G protein-biased ligand at the μ-opioid receptor is potently analgesic with reduced gastrointestinal and respiratory dysfunction compared with morphines. J Pharmacol Exp Ther 344(3):708–717. https://doi.org/10.1124/jpet.112.201616
Kruegel AC et al (2016) Synthetic and receptor signaling explorations of the mitragyna alkaloids: mitragynine as an atypical molecular framework for opioid receptor modulators. J Am Chem Soc 138(21):6754–6764. https://doi.org/10.1021/jacs.6b00360
Kennedy NM et al (2018) Optimization of a series of Mu Opioid Receptor (MOR) agonists with high G protein signaling bias. J Med Chem. https://doi.org/10.1021/acs.jmedchem.8b01136
Koehl A et al (2018) Structure of the µ-opioid receptor–G i protein complex. Nature. https://doi.org/10.1038/s41586-018-0219-7
“Chemical Computing Group (CCG) | Research.” https://www.chemcomp.com/Research-Citing_MOE.htm (Accessed Jun. 29, 2021).
Bartuzi D, Kaczor AA, Matosiuk D (2015) Activation and allosteric modulation of human μ opioid receptor in molecular dynamics. J Chem Inf Model 55(11):2421–2434. https://doi.org/10.1021/acs.jcim.5b00280
Jo S, Kim T, Iyer VG, Im W (2008) CHARMM-GUI: A web-based graphical user interface for CHARMM. J Comput Chem 29(11):1859–1865. https://doi.org/10.1002/jcc.20945
Salomon-Ferrer R, Case DA, Walker RC (2013) An overview of the Amber biomolecular simulation package. WIREs Comput Mol Sci 3(2):198–210. https://doi.org/10.1002/wcms.1121
Götz AW, Williamson MJ, Xu D, Poole D, Le Grand S, Walker RC (2012) Routine microsecond molecular dynamics simulations with AMBER on GPUs. 1. Generalized born. J Chem Theory Comput 8(5):1542–1555. https://doi.org/10.1021/ct200909j
Gatica EA, Cavasotto CN (2012) Ligand and decoy sets for docking to G protein-coupled receptors. J Chem Inf Model 52(1):1–6. https://doi.org/10.1021/ci200412p
Mansour A et al (2002) Key residues defining the μ-opioid receptor binding pocket: a site-directed mutagenesis study. J Neurochem 68(1):344–353. https://doi.org/10.1046/j.1471-4159.1997.68010344.x
Manglik A et al (2012) Crystal structure of the mu-opioid receptor bound to a morphinan antagonist. Nature. https://doi.org/10.1038/nature10954
Yuan S, Vogel H, Filipek S (2013) The role of water and sodium ions in the activation of the μ-opioid receptor. Angew Chem Int Ed 52(38):10112–10115. https://doi.org/10.1002/anie.201302244
Marmolejo-Valencia AF, Madariaga-Mazón A, Martinez-Mayorga K (2021) Bias-inducing allosteric binding site in mu-opioid receptor signaling. SN Appl Sci. https://doi.org/10.1007/s42452-021-04505-8
Pasternak GW (2014) Opioids and their receptors: Are we there yet? Neuropharmacology. https://doi.org/10.1016/j.neuropharm.2013.03.039
Yudin Y, Rohacs T (2019) The G-protein-biased agents PZM21 and TRV130 are partial agonists of μ-opioid receptor-mediated signalling to ion channels. Br J Pharmacol. https://doi.org/10.1111/bph.14702
Harding WW et al (2005) Neoclerodane diterpenes as a novel scaffold for μ opioid receptor ligands. J Med Chem 48(15):4765–4771. https://doi.org/10.1021/jm048963m
Wang Y et al (2008) 2-Methoxymethyl-salvinorin B is a potent κ opioid receptor agonist with longer lasting action in vivo than salvinorin A. J Pharmacol Exp Ther 324(3):1073–1083. https://doi.org/10.1124/jpet.107.132142
Pedersen MH, Pham J, Mancebo H, Inoue A, Asher WB, Javitch JA (2021) A novel luminescence-based β-arrestin recruitment assay for unmodified receptors. J Biol Chem. https://doi.org/10.1016/j.jbc.2021.100503
Ortega A, Blount JF, Manchand PS (1982) Salvinorin, a new trans-neoclerodane diterpene from Salvia divinorum (Labiatae). J Chem Soc Perkin. https://doi.org/10.1039/p19820002505
Hernández-Alvarado RB, Madariaga-Mazón A, Ortega A, Martinez-Mayorga K (2020) DARK classics in chemical neuroscience: Salvinorin A. ACS Chem Neurosci 11(23):3979–3992. https://doi.org/10.1021/acschemneuro.0c00608
Groer CE et al (2007) An opioid agonist that does not induce μ-opioid receptor - Arrestin interactions or receptor internalization. Mol Pharmacol 71(2):549–557. https://doi.org/10.1124/mol.106.028258
Kane BE, Nieto MJ, McCurdy CR, Ferguson DM (2006) A unique binding epitope for salvinorin A, a non-nitrogenous kappa opioid receptor agonist. FEBS J 273(9):1966–1974. https://doi.org/10.1111/j.1742-4658.2006.05212.x
Schneider S, Provasi D, Filizola M (2016) How oliceridine (TRV-130) binds and stabilizes a μ-opioid receptor conformational state that selectively triggers G protein signaling pathways. Biochemistry 55(46):6456–6466. https://doi.org/10.1021/acs.biochem.6b00948
Lipiński PFJ, Jarończyk M, Dobrowolski JC, Sadlej J (2019) Molecular dynamics of fentanyl bound to μ-opioid receptor. J Mol Model 25(5):1–17. https://doi.org/10.1007/s00894-019-3999-2
Hulme EC (2013) GPCR activation: A mutagenic spotlight on crystal structures. Trends Pharmacol Sci 34(1):67–84. https://doi.org/10.1016/j.tips.2012.11.002
Kaiser A, Hempel C, Wanka L, Schubert M, Hamm HE, Beck-Sickinger AG (2018) G protein preassembly rescues efficacy of W 6.48 toggle mutations in neuropeptide Y 2 receptor. Mol Pharmacol 93(4):387–401. https://doi.org/10.1124/mol.117.110544
Schmid CL et al (2017) Bias factor and therapeutic window correlate to predict safer opioid analgesics. Cell. https://doi.org/10.1016/j.cell.2017.10.035
Kenakin T (2011) Functional selectivity and biased receptor signaling. J Pharmacol Exp Ther 336(2):296–302. https://doi.org/10.1124/jpet.110.173948
Black JW, Leff P (1983) Operational models of pharmacological agonism. Proc R Soc Lond B Biol Sci 220(1219):141–162. https://doi.org/10.1098/rspb.1983.0093
Harrison C, Traynor JR (2003) The [35S]GTPγS binding assay: Approaches and applications in pharmacology. Life Sci 74(4):489–508. https://doi.org/10.1016/j.lfs.2003.07.005
Remy I, Michnick SW (2007) Application of protein-fragment complementation assays in cell biology. Biotechniques 42(2):137–145. https://doi.org/10.2144/000112396
Pil J, Tytgat J (2003) Serine 329 of the μ-opioid receptor interacts differently with agonists. J Pharmacol Exp Ther 304(3):924–930. https://doi.org/10.1124/jpet.102.040113
Granier S et al (2012) Structure of the δ-opioid receptor bound to naltrindole. Nat Lond 485(7398):400–404
Che T et al (2018) Structure of the nanobody-stabilized active state of the kappa opioid receptor. Cell 172(1–2):55–67. https://doi.org/10.1016/j.cell.2017.12.011
Kapoor A, Martinez-Rosell G, Provasi D, De Fabritiis G, Filizola M (2017) Dynamic and kinetic elements of μ-opioid receptor functional selectivity. Sci Rep 7(1):1–15. https://doi.org/10.1038/s41598-017-11483-8
Sader S, Anant K, Wu C (2018) To probe interaction of morphine and IBNtxA with 7TM and 6TM variants of the human μ-opioid receptor using all-atom molecular dynamics simulations with an explicit membrane. Phys Chem Chem Phys 20(3):1724–1741. https://doi.org/10.1039/c7cp06745c
Kobilka BK, Deupi X (2007) Conformational complexity of G-protein-coupled receptors. Trends Pharmacol Sci 28(8):397–406. https://doi.org/10.1016/j.tips.2007.06.003
Vaidehi N, Kenakin T (2010) The role of conformational ensembles of seven transmembrane receptors in functional selectivity. Curr Opin Pharmacol 10(6):775–781. https://doi.org/10.1016/j.coph.2010.09.004
Wingler LM et al (2019) Angiotensin analogs with divergent bias stabilize distinct receptor conformations. Cell 176(3):468–478. https://doi.org/10.1016/j.cell.2018.12.005
Wolf A, Kirschner KN (2013) Principal component and clustering analysis on molecular dynamics data of the ribosomal L11·23S subdomain. J Mol Model 19(2):539–549. https://doi.org/10.1007/s00894-012-1563-4
Granier S, Kobilka B (2012) A new era of GPCR structural and chemical biology. Nat Chem Biol 8(8):670–673. https://doi.org/10.1038/nchembio.1025
A. Malvezzi, L. Rezende, M. A. Izidoro, M. Sedenho, L. Juliano and A. T.-do Amaral, “Uncovering false positives on a virtual screening search for cruzain inhibitors” Bioorg Med Chem Lett, 2008, doi: https://doi.org/10.1016/j.bmcl.2007.10.068.
Jorgensen WL (2004) The many roles of computation in drug discovery. Science 303(5665):1813–1818. https://doi.org/10.1126/science.1096361
Acknowledgements
This work was supported by Instituto de Química. The computations were supported by the Dirección General de Cómputo y de Tecnologías de Información y Comunicación (DGTIC)-UNAM, providing Miztli computer resources under grant LANCAD-UNAM-DGTIC-367. RBH-A acknowledges CONACyT (957374) for scholarship, FC-V acknowledges a scholarship from QUIBIC Group, UNAM. KM-M thanks DGAPA-PASPA and AN thanks NIDA (Grant: R33DA044425) for financial support. Authors thank Dr. Alan Grossfield and Dr. Rogelio Rodríguez-Sotres for insightful discussions. To the developers of ChemAxon, DataWarrior, Amber MD package, VMD/NAMD for kindly providing academic licenses of their software.
Funding
DGAPA, UNAM, Programa de Apoyos para la Superación del Personal Académico (PASPA).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
Not applicable.
Consent for publication
Not applicable
Consent to participate
Not applicable
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Hernández-Alvarado, R.B., Madariaga-Mazón, A., Cosme-Vela, F. et al. Encoding mu-opioid receptor biased agonism with interaction fingerprints. J Comput Aided Mol Des 35, 1081–1093 (2021). https://doi.org/10.1007/s10822-021-00422-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10822-021-00422-5