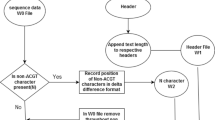
Abstract
With the initial completion of Human Genome Project, the post-genomic era is coming. Although the genome map of human has been decoded, the roles that each segment of sequences acts are not totally discovered. On the other hand, with the rapid expansion of sequence information, the issues of data compilation and data storage are increasingly important. In this paper, a “Human genome database system” is designed and implemented in National Taiwan University Hospital (NTUH). By accessing this system, the doctors can store and manage the experimental sequence data. The achievement of this system is that it integrates the modules of sequence alignment and data compression. By embedding with the NCBI alignment program—blastall [1], it automatically aligns the uploaded sequences and searches for the corresponding genomic positions. Besides, the system encodes the differences between sequences, effectively compresses them and decreases the demand of storage spaces by the compression ratio at 12.28. At the same time, it offers a variety of query methods. Users can quickly access the interesting data by inputting the keywords of specimen number, GI and sequence position, etc. The electronic health record (EHR) in Health Information System (HIS) of NTUH is also combined in this system and the doctors can utilize the valuable information to figure out the relation between the diseases and genes. With this system, a genetic personal healthcare environment will be established in the future.














Similar content being viewed by others
References
Blastall. http://www.ncbi.nlm.nih.gov/staff/tao/URLAPI/blastall/.
DNA sequencing. http://genomics.org/index.php/DNA_sequencing.
Sanger, F., Air, G. M., Barrell, B. G., Brown, N. L., Coulson, A. R., Fiddes, C. A., Hutchison, C. A., Slocombe, P. M., and Smith, M., Nucleotide sequence of bacteriophage phi X174 DNA. Nature 265(5596):687–695, 1977.
Shendure, J., and Ji, H., Next-generation DNA sequencing. Nat. Biotechnol. 26(10):1135–1145, 2008.
Human Genome Project HomePage. http://hgph.com/index.htm.
Human Genome Project Information. http://www.ornl.gov/sci/techresources/Human_Genome/home.shtml.
McGinnis, S., and Madden, T. L., BLAST: at the core of a powerful and diverse set of sequence analysis tools. Nucleic Acids Res. 32:20–25, 2004.
Benson, D. A., Boguski, M. S., Lipman, D. J., Ostell, J., Ouellette, B. F., Rapp, B. A., and Wheeler, D. L., GenBank. Nucleic Acids Res. 27(1):12–17, 1999.
Hubbard, T., Barker, D., Birney, E., Cameron, G., Chen, Y., Clark, L., Cox, T., Cuff, J., Curwen, V., Down, T., Durbin, R., Eyras, E., Gilbert, J., Hammond, M., Huminiecki, L., Kasprzyk, A., Lehvaslaiho, H., Lijnzaad, P., Melsopp, C., Mongin, E., Pettett, R., Pococok, M., Potter, S., Rust, A., Schmidt, E., Searles, S., Slater, G., Smith, J., Spooner, W., Stabenau, A., Stalker, J., Stupka, E., Ureta-Vidal, A., Vastrk, I., and Clamp, M., The Ensembl genome database project. Nucleic Acids Res. 30(1):38–41, 2002.
Tateno, Y., Miyazaki, S., Ota, M., Sugawara, M., and Gojobori, T., DNA Data Bank of Japan (DDBJ) in collaboration with mass sequencing teams. Nucleic Acids Res. 28(1):24–26, 2000.
Blake, J. A., Richardson, J. E., Bult, C. J., Kadin, J. A., and Eppig, J. P., MGD: the Mouse Genome Database. Nucleic Acids Res. 31(1):193–195, 2003.
Maruyama, Y., Wakamatsu, A., Kawamura, Y., Kimura, K., Yamamoto, J., Nishikawa, T., Kisu, Y., Sugano, S., Goshima, N., Isogai, T., and Nomura, N., Human Gene and Protein Database (HGPD): a novel database presenting a large quantity of experiment-based results in human proteomics. Nucleic Acids Res. 37:762–766, 2009.
Pruitt, K. D., Tatusova, T., and Maglott, D. R., NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 35:61–65, 2006.
NCBI Reference Sequence (RefSeq). http://www.ncbi.nlm.nih.gov/RefSeq/.
BLAST Program Selection Guide. http://blast.ncbi.nlm.nih.gov/blast/producttable.shtml.
MegaBLAST Search. http://www.ncbi.nlm.nih.gov/blast/megablast.shtml.
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipmen, D. J., Basic local alignment search tool. J. Mol. Biol. 215(3):403–410, 1990.
Grumbach, S. and Tahi, F., Compression of DNA sequences. Data Compression Conference. (pp. 340–350). IEEE Computer Society Press, 1993.
Chen, X., Kwong, S., and Li, M., A compression algorithm for DNA sequences and its applications in genome comparison. The 10th Workshop on Genome Informatics. (pp. 51–61). Genome Informatics Press, 1999
Chen, X., Li, M., Ma, B., and Tromp, J., DNACompress: fast and effective DNA sequence compression. Bioinformatics 18(12):1696–1698, 2002.
Ma, B., Tromp, J., and Li, M., PatternHunter: faster and more sensitive homology search. Bioinformatics 18(3):440–445, 2002.
Behzadi, B., and Fessant, F. L., DNA compression challenge revisited: a dynamic programming approach. Lect. Notes Comput. Sci. 3537:190–200, 2005.
Jurka, J., Kapitonov, V. V., Pavlice, A., Klonowski, P., Kohany, O., and Walichiewicz, J., Repbase update, a database of eukaryotic repetitive elements. Cytogenet. Genome Res. 110(1–4):462–467, 2005.
FASTA format description. http://www.ncbi.nlm.nih.gov/blast/fasta.shtml.
Apostolico, A., and Fraenkel, A., Robust transmission of unbounded strings using Fibonacci representations. IEEE Trans. Inf. Theory 33(2):238–245, 1987.
Elias, P., Universal codeword sets and representations of the integers. IEEE Trans. Inf. Theory 21(2):194–203, 1975.
Li, R., Li, Y., Kristiansen, K., and Wang, J., SOAP: short oligonucleotide alignment program. Bioinformatics 24(5):713–714, 2008.
Genetic Information Nondiscrimination Act of 2008 (Public Law 110–233).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Chen, WH., Lu, YW., Lai, F. et al. Integrating Human Genome Database into Electronic Health Record with Sequence Alignment and Compression Mechanism. J Med Syst 36, 2587–2597 (2012). https://doi.org/10.1007/s10916-011-9731-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10916-011-9731-0