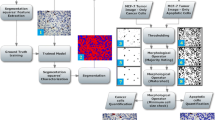
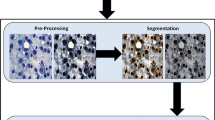
Abstract
Based on the Nottingham criteria, the number of mitosis cells in histopathological slides is an important factor in diagnosis and grading of breast cancer. For manual grading of mitosis cells, histopathology slides of the tissue are examined by pathologists at 40× magnification for each patient. This task is very difficult and time-consuming even for experts. In this paper, a fully automated method is presented for accurate detection of mitosis cells in histopathology slide images. First a method based on maximum-likelihood is employed for segmentation and extraction of mitosis cell. Then a novel Maximized Inter-class Weighted Mean (MIWM) method is proposed that aims at reducing the number of extracted non-mitosis candidates that results in reducing the false positive mitosis detection rate. Finally, segmented candidates are classified into mitosis and non-mitosis classes by using a support vector machine (SVM) classifier. Experimental results demonstrate a significant improvement in accuracy of mitosis cells detection in different grades of breast cancer histopathological images.













Similar content being viewed by others
References
Frierson, J H. F., Interobserver reproducibility of the Nottingham modification of the bloom and richardson histologic grading scheme for infiltrating ductal carcinoma. Am. J. Clin. Pathol. 195–198, 1995
He, L., Long, L.R, Antani, S. and Thoma, G.R., Histology image analysis for carcinoma detection and grading. Comput. Methods Prog. Biomed. 538–556, 2012.
Ciresan., D. C. Mitosis detection in breast cancer histology images with deep neural networks, in: MICCAI 2013. Springer, 411–418, 2013.
Nateghi, R., Danyali, H., Helfroush, M. S. and Tashk, A., Intelligent cad system for automatic detection of mitosis cells from breast cancer histology slide images based on teaching- learning based optimization, Comput. Biol. J, 2014.
Khan, A.M. El-Daly H. and Rajpoot., N. M. A gamma-gaussian mixture model for detection of mitosis cells in breast cancer histopathology images. In: Pattern Recognition (ICPR), 2012, 21st International Conference on. IEEE, 149–152, 2012.
Sommer, C. Fiaschi, L. Hamprecht F. A. and Gerlich., D. W. Learning-based mitosis cell detection in histopathological images. In: Pattern Recognition (ICPR), International Conference on. IEEE, 2306–2309, 2012.
Irshad, H., Jalali, S., Roux, L., Racoceanu, D., Hwee, L. J., Le Naour, G., and Capron, F., Automated mitosis detection using texture, sift features and HMAX biologically inspired approach. J. pathol. Inf., 2013.
Paul A. and Mukherjee., D. P. Mitosis detection for invasive breast cancer grading in histopathological images. Image Processing. IEEE Transaction on, 4041–4054, 2015.
Tashk, A., Helfroush. M. S., Danyali, H. and Akbarzadeh Jahromi, M., Automatic detection of breast cancer mitosis cells based on the combination of textural, statistical and innovative mathematical features. Appl. Math. Model., 6165–6182, 2015.
Chen, H. Dou, Q. Wang, X. Qin J. and Heng., P. Mitosis detection in breast cancer histology images via deep cascaded networks. In: Proceedings of the Thirtieth AAAI Conference on Artificial Intelligence, 1160–1166, 2016.
Roullier, V., Lezoray, O., Ta, V.T., and Elmoataz, A., Multi-resolution graph-based analysis of histopathological whole slide images, Application to mitosis cell extraction and visualization. Comput. Med. Imaging Graph., 603–615, 2011.
Tek, F. B., Mitosis detection using generic features and an ensemble of cascade adaboosts. J. Pathol. Inf., 2013.
Lu, C. and Mandal, M.., Towards automatic mitosis cells detection and segmentation in multi-spectral histopathological images. IEEE J. Biomed. and Health Inform., 594–605, 2013.
Irshad, H. Gouaillard, A. Roux L. and Racoceanu., D. Spectral band selection for mitosis detection in histopathology. In: Biomedical Imaging (ISBI), 11st International Symposium on. IEEE, 1279–1282, 2014.
Irshad, H., Gouaillard, A., Roux, L. and Racoceanu, D., Multispectral band selection and spatial characterization, application to mitosis detection in breast cancer histopathology. Comput. Med. Imaging Graph, 390–402, 2014.
Macenko, M. Niethammer, M. Marron, J. S. Borland, D. Woosley, JT Xiaojun, G. Schmitt C. and Thomas., NE. A method for normalizing histology slides for quantitative analysis. In: Biomedical Imaging (ISBI), International Symposium on. IEEE, 1107–1110, 2009.
Khan, A. M. Rajpoot, N. Treanor, D. Magee., D. A nonlinear mapping approach to stain normalization in digital histopathology images using image-specific color deconvolution. Biomedical Engineering, IEEE Transactions on, 1729–1738, 2014.
Yang., X. Nuclei segmentation using marker-controlled watershed, tracking using mean-shift, and kalman filter in time-lapse microscopy. Circuits and Systems I, Regular Papers, IEEE Trans. on, 2405–2414, 2006.
Al-Kofahi., Y. Improved automatic detection and segmentation of cell nuclei in histopathology images. Biomedical Engineering, IEEE Transactions on, 841–852, 2010.
Mukherjee, D. P. Ray, N. and Acton., S. T. Level set analysis for leukocyte detection and tracking. Image Processing, IEEE Transactions on, 562–572, 2004.
Plissiti, M. E. Nikou., C. Overlapping cell nuclei segmentation using a spatially adaptive active physical model. Image Processing, IEEE Transactions on, 4568–4580, 2012.
Otsu., N. A threshold selection method from gray-level histograms. Systems, Man and Cybernetics, IEEE Transaction on, 62–66, 1979.
Nagase, A. Takahashi M. and Nakano., M. Automatic calculation and visualization of nuclear density in whole slide images of hepatic histological sections. Bio-Med. Mater. Eng., 1335–1344, 2015.
Karagiannidis G. K. and Lioumpas., A. S. An improved approximation for the Gaussian Q-function. IEEE Communications Letters, 644–646, 2007.
Haralick, R. M. Shanmygam K. and Dinstein., I. Textural features for image classification. Systems, Man and Cybernetics, IEEE Transaction on, 610–621, 1973.
Tang., X. Texture information in run-length matrices. Image Processing, IEEE Transactions on, 1602–1609, 1998.
Guo., Z. A complete modeling of local binary pattern operator for texture classification. Image Processing, IEEE Transactions on, 1657–1663, 2010.
Cortes, C. Vapnik., V. Support-vector networks. Machine Learning Springer, 273–297, 1995.
Available as on 10.06.2014. [Online]. Available, http://mitos-atypia-14.grand-challenge.org/dataset/
Available as on 5.02.2013. [Online]. Available: http://amida13.isi.uu.nl/?q=node/62
Available as on 8.01.2012. [Online]. Available: http://ludo17.free.fr/mitos_2012/dataset.html
Perona, P. and Malik, J.., Scale-space and edge detection using anisotropic diffusion. Pattern Analysis and Machine Intelligence, IEEE Transactions on, 629–639, 1990.
Xu., L. Image smoothing via l 0 gradient minimization. ACM Trans. Graph. 174, 2011.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
Author A declares that he has no conflict of interest. Author B declares that he has no conflict of interest. Author C declares that he has no conflict of interest.
Ethical Approval
This article does not contain any studies with human participants performed by any of the authors.
Additional information
This article is part of the Topical Collection on Image & Signal Processing
Rights and permissions
About this article
Cite this article
Nateghi, R., Danyali, H. & Helfroush, M.S. Maximized Inter-Class Weighted Mean for Fast and Accurate Mitosis Cells Detection in Breast Cancer Histopathology Images. J Med Syst 41, 146 (2017). https://doi.org/10.1007/s10916-017-0773-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10916-017-0773-9