Abstract
This paper aims at developing an automatic pathological brain detection system (PBDS) to assist radiologists in identifying brain diseases correctly in less time. Magnetic resonance imaging (MRI) has the potential to provide better information about the brain soft tissues and hence MR images have been incorporated in the proposed system. Fifty largest coefficients are selected from each sub-band of a level-5 fast discrete curvelet transform (FDCT) to serve as a feature set for each image. To reduce the size of the feature set, principal component analysis (PCA) has been harnessed. Subsequently, least squares SVM (LS-SVM) with three different kernels are utilized to segregate the images as healthy or pathological. The proposed system has been validated on three benchmark datasets and a 10 ×k-fold stratified cross validation (SCV) test has been performed. It indicates that the proposed system “FDCT + PCA + LS-SVM + RBF” achieves better performance than not only two other systems having linear and polynomial kernel but also 22 existing methods. In addition, the suggested system requires only six features which are computationally economical for a practical use.









Similar content being viewed by others
References
Bishop CM (2006) Pattern recognition and machine learning. Springer, New York
Candès EJ (2003) What is... a curvelet? Not Amer Math Soc 50(11):1402–1403
Candès EJ, Donoho DL (1999) Ridgelets: A key to higher-dimensional intermittency? Philosophical transactions of the royal society of london a: mathematical. Phys Eng Sci 357(1760):2495–2509
Candès EJ, Donoho DL (2000) Curvelets- a surprisingly effective nonadaptive representation for objects with edges. Vanderbilt University Press, Nashville
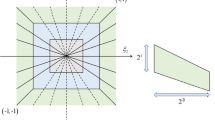
Candes E, Demanet L, Donoho D, Ying L (2006) Fast discrete curvelet transforms. Multiscale Model Simul 5(3):861–899
Chaplot S, Patnaik LM, Jagannathan NR (2006) Classification of magnetic resonance brain images using wavelets as input to support vector machine and neural network. Biomed Signal Process Control 1(1):86–92
Chen Y, Shi L, Feng Q, Yang J, Shu H, Luo L, Coatrieux JL, Chen W (2014) Artifact suppressed dictionary learning for low-dose CT image processing. IEEE Trans Med Imag 33(12):2271–2292
Das S, Chowdhury M, Kundu K (2013) Brain MR image classification using multiscale geometric analysis of ripplet. Progress Electromagn Res 137:1–17
Do MN, Vetterli M (2003) The finite ridgelet transform for image representation. IEEE Trans Image Process 12(1):16–28
Duda RO, Hart PE, Stork DG (2012) Pattern classification. Wiley, New York
El-Dahshan ESA, Honsy T, Salem ABM (2010) Hybrid intelligent techniques for MRI brain images classification. Digit Signal Process 20(2):433–441
El-Dahshan EA, Mohsen HM, Revett K, Salem ABM (2014) Computer-aided diagnosis of human brain tumor through MRI: a survey and a new algorithm. Expert Syst Appl 41(11):5526–5545
Johnson KA, Becker JA The whole brain atlas. http://www.med.harvard.edu/AANLIB/
Maitra M, Chatterjee A (2006) A Slantlet transform based intelligent system for magnetic resonance brain image classification. Biomed Signal Process Control 1 (4):299–306
Nayak DR, Dash R, Majhi B (2015) Classification of brain MR images using discrete wavelet transform and random forests. In: Fifth National conference on computer vision, pattern recognition, image processing and graphics (NCVPRIPG). IEEE, pp 1–4
Nayak DR, Dash R, Majhi B (2016) Brain MR image classification using two-dimensional discrete wavelet transform and AdaBoost with random forests. Neurocomputing 177:188–197
Nayak DR, Dash R, Majhi B, Mohammed J (2016) Non-linear cellular automata based edge detector for optical character images. Simulation:1–11
Pisano ED, Zong S, Hemminger BM, DeLuca M, Johnston RE, Muller K, Braeuning MP, Pizer SM (1998) Contrast limited adaptive histogram equalization image processing to improve the detection of simulated spiculations in dense mammograms. J Digit Imag 11(4):193–200
Pizer SM, Johnston R E, Ericksen JP, Yankaskas BC, Muller KE (1990) Contrast-limited adaptive histogram equalization: speed and effectiveness. In: Proceedings of the first conference on visualization in biomedical computing. IEEE, pp 337–345
Reza AM (2004) Realization of the contrast limited adaptive histogram equalization (CLAHE) for real-time image enhancement. J VLSI Signal Process Syst Signal Image Vid Technol 38(1):35–44
Saritha M, Joseph KP, Mathew AT (2013) Classification of MRI brain images using combined wavelet entropy based spider web plots and probabilistic neural network. Pattern Recog Lett 34(16):2151–2156
Suykens JAK, Vandewalle J (1999) Least squares support vector machine classifiers. Neural Process Lett 9(3):293–300
Wang S, Zhang Y, Dong Z, Du S, Ji G, Yan J, Yang J, Wang Q, Feng C, Phillips P (2015) Feed-forward neural network optimized by hybridization of PSO and ABC for abnormal brain detection. Int J Imag Syst Technol 25(2):153–164
Wang S, Zhang Y, Yang X, Sun P, Dong Z, Liu A, Yuan TF (2015) Pathological brain detection by a novel image feature—fractional Fourier entropy. Entropy 17(12):8278–8296
Wang S, Lu S, Dong Z, Yang J, Yang M, Zhang Y (2016) Dual-tree complex wavelet transform and twin support vector machine for pathological brain detection. Appl Sci 6(6):169
Wang S, Phillips P, Yang J, Sun P, Zhang Y (2016) Magnetic resonance brain classification by a novel binary particle swarm optimization with mutation and time-varying acceleration coefficients. Biomed Eng/Biomedizinische Technik:1–10
Westbrook C (2014) Handbook of MRI technique. Wiley, Oxford
Yang G, Zhang Y, Yang J, Ji G, Dong Z, Wang S, Feng C, Wang Q (2015) Automated classification of brain images using wavelet-energy and biogeography-based optimization. Multimed Tools Appl:1–17
Zhang Y, Wu L (2012) An MR brain images classifier via principal component analysis and kernel support vector machine. Progress Electromagn Res 130:369–388
Zhang Y, Wang S, Wu L (2010) A novel method for magnetic resonance brain image classification based on adaptive chaotic PSO. Progress Electromagn Res 109:325–343
Zhang Y, Dong Z, Wu L, Wang S (2011) A hybrid method for MRI brain image classification. Expert Syst Appl 38(8):10,049–10,053
Zhang Y, Wu L, Wang S (2011) Magnetic resonance brain image classification by an improved artificial bee colony algorithm. Progress Electromagn Res 116:65–79
Zhang Y, Wang S, Ji G, Dong Z (2013) An MR brain images classifier system via particle swarm optimization and kernel support vector machine. Sci World J 2013:1–9
Zhang G, Wang Q, Feng C, Lee E, Ji G, Wang S, Zhang Y, Yan J (2015) Automated classification of brain MR images using wavelet-energy and support vector machines. In: 2015 International conference on mechatronics, electronic, industrial and control engineering (MEIC-15), pp 683–686
Zhang Y, Dong Z, Liu A, Wang S, Ji G, Zhang Z, Yang J (2015) Magnetic resonance brain image classification via stationary wavelet transform and generalized eigenvalue proximal support vector machine. J Med Imag Health Inf 5 (7):1395–1403
Zhang YD, Chen S, Wang SH, Yang JF, Phillips P (2015) Magnetic resonance brain image classification based on weighted-type fractional Fourier transform and nonparallel support vector machine. Int J Imag Syst Technol 25(4):317–327
Zhang Y, Wang S, Sun P, Phillips P (2015) Pathological brain detection based on wavelet entropy and Hu moment invariants. Bio-Med Mater Eng 26(s1):S1283–S1290
Zhang Y, Wang S, Dong Z, Phillip P, Ji G, Yang J (2015) Pathological brain detection in magnetic resonance imaging scanning by wavelet entropy and hybridization of biogeography-based optimization and particle swarm optimization. Progress Electromagn Res 152:41–58
Zhang YD, Wang SH, Yang XJ, Dong ZC, Liu G, Phillips P, Yuan TF (2015) Pathological brain detection in MRI scanning by wavelet packet Tsallis entropy and fuzzy support vector machine. SpringerPlus 4(1):1–16
Zhang Y, Dong Z, Wang S, Ji G, Yang J (2015) Preclinical diagnosis of magnetic resonance (MR) brain images via discrete wavelet packet transform with Tsallis entropy and generalized eigenvalue proximal support vector machine (GEPSVM). Entropy 17(4):1795–1813
Zhang Y, Sun Y, Phillips P, Liu G, Zhou X, Wang S (2016) A multilayer perceptron based smart pathological brain detection system by fractional Fourier entropy. J Med Syst 40(7):1–11
Zhou X, Wang S, Xu W, Ji G, Phillips P, Sun P, Zhang Y (2015) Detection of pathological brain in MRI scanning based on wavelet-entropy and naive Bayes classifier. In: Bioinformatics and biomedical engineering, pp 201–209
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Nayak, D.R., Dash, R. & Majhi, B. Pathological brain detection using curvelet features and least squares SVM. Multimed Tools Appl 77, 3833–3856 (2018). https://doi.org/10.1007/s11042-016-4171-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11042-016-4171-y