Abstract
In the last decade, supervoxels have become a useful mid-level representation of volumetric medical images such as MRIs and CT scans. Several methods were suggested to produce uniform supervoxels, yet little has been done to generate content-sensitive over-segmentations. This is particularly beneficial to 3D medical image analysis, where sizes of anatomical structures vary largely. In this paper, we propose AdaSLIC as an adaptive supervoxel generation technique that applies to volumetric medical images. In small structures, it generates tiny supervoxels to capture the details of the image. Meanwhile, it partitions large structures into bigger supervoxels, hence leading to a sparse description. The proposed technique is an extension of the Simple Linear Iterative Clustering (SLIC) algorithm. Rather than using a regular sampling to initiate supervoxel centers, a content-sensitive initialization is performed using a Poisson-disk sampling algorithm (PDS). It relies on a map of distances to the main image contours. The size of each supervoxel depends on the distance of its center to the closest image contour. We compare our algorithm to the SLIC algorithm as well as to an extension of the DBSCAN algorithm (Density-Based Spatial Clustering of Applications with Noise). Two datasets are used for this purpose: knee MRIs and cardiovascular magnetic resonance (CMR) images. We use different metrics to assess the quality of the generated over-segmentations. Experimental results show that our algorithm achieves comparable or better boundary adherence than the state of the art algorithms while producing compact and adaptive supervoxels.
Similar content being viewed by others
1 Introduction
The large size of 3D medical images has always been an important limitation for an efficient analysis. As a result, a significant number of methods have recently shifted to the use of supervoxels as a mid-level representation. Images are over-segmented into meaningful regions by grouping voxels with similar features and belonging to the same anatomical structures. Adopting this mid-level representation reduces the complexity of several image processing tasks. For instance, supervoxels have been used for image registration [8, 18, 22, 36, 44], tumor delineation [20, 27], segmentation [1, 2, 45] and classification [11, 31].
A useful supervoxel representation has to be computationally efficient [42] and has to exhibit some properties. In this work, out of the widely studied properties, we focus on boundary adherence [1] and compactness [40]. Boundary adherence is arguably one of the most important properties. It reflects how supervoxel boundaries fit to object borders and requires that each supervoxel overlaps with a single object/structure in the image. As for the compactness, it expresses how smooth and regular is the shape of supervoxels.
Recently, an interest has been given to the sensitivity of the over-segmentation to image content. It implies adapting the size of superpixels to the image underlying structures: tiny superpixels are generated within small structures and larger ones partition bigger structures. Adaptive superpixels lead to a more compressed representation while preserving the adherence to image boundaries. Consequently, this mid-level representation will further reduce the complexity of subsequent image processing tasks, particularly for large size data, such as volumetric medical images [11, 27, 45]. For instance, in recent graph based segmentation methods [18, 29, 44], supervoxels are used as graph nodes. The computation cost of these algorithms is highly affected by the number of supervoxels. Hence, an adaptive over-segmentation which reduces this number is highly recommended. The same impact applies to Random Forests based segmentation and registration where supervoxels are used as a mid-level representation [8, 21, 31]. Moreover, adaptive supervoxels are beneficial to image compression [24]. On the one hand, small segments preserve important features in dense regions and therefore decrease the compression error. On the other hand, large segments in sparse regions increase the compression capability of the representation.
Adaptive supervoxels have been mainly generated for 2D cases. The main contribution of this paper is to extend the content-sensitivity property to volumetric images such as MR and CMR images. To address this problem, we propose an adaptive supervoxel generation algorithm, that we name AdaSLIC. It is based on an extension of the SLIC [1] algorithm. This classical superpixels/voxels generation technique is initiated by a regularly sampled seeds (cluster centers). Pixels/voxels are then iteratively clustered according to their proximity and similarity to the cluster centers. The SLIC algorithm generates regular-sized superpixels/voxels.
To produce an adaptive over-segmentation, we first define a density function based on a distance map to the main contours of the image. Then, we perform a PDS algorithm [13] to automatically sample the seeds according to the density function. This step is followed by an adaptive clustering where we the sizes of supervoxels are deduced from the density function.
Experimental results, carried out on two sets of MR images, show that our algorithm achieves a good boundary adherence compared to the state of the art algorithms while considerably reducing the number of supervoxels.
The remainder of this paper is organized as follows. In Section 2, we review the principal techniques for superpixel and supervoxel generation. Then, in Section 3 we briefly describe the SLIC and PDS algorithms as preliminaries to our work. In Section 4, we present the AdaSLIC technique. The experimental results and the evaluation of the method are detailed in Section 5. We discuss the limitations of our method and the possible improvements in Section 6. Finally, we conclude in Section 7.
2 Related work
Several superpixel and supervoxel generation algorithms have been introduced in the last decades. In this section, we first review the main approaches. Second, we focus on content-sensitive techniques. Then, we highlight the contribution brought by our method in this context.
Superpixel generation algorithms can be roughly divided into two main groups, namely graph based methods and clustering based methods.
Graph-based methods employ a graph representation of the image where nodes usually correspond to image pixels and edges weigh the similarity between neighboring nodes. Superpixels are generated by minimizing an energy function using algorithms such as graph cut.
Ren and Malik [38] introduced an over-segmentation algorithm based on the Normalized Cuts (Ncuts) [43]. The produced superpixels exhibit a regular shape. However, they present a low boundary adherence and their generation is achieved at a high computational cost.
In [14], Felzenszwalb and Huttenlocher group pixels/regions exhibiting similar appearance. Each superpixel is the minimum spanning tree of the constituent pixels. Even though the method shows a high boundary adherence and requires a low computation time, it generates superpixels with irregular sizes and shapes. Recently, this method was extended to improve the boundary adherence by generating a hierarchical over-segmentation [16]. However, similar to the original one-level algorithm, the produced superpixels/voxels exhibit a low compactness. Moore et al. [33] introduced the superpixel lattices to generate regular superpixels with a low computational cost. The algorithm relies on a boundary map to generate grid-like superpixels. It sequentially cuts the image by searching for optimal paths within a vertical and a horizontal strips.
In [41, 47], a compromise between shape regularity and boundary adherence is achieved. In [47], the image is initially covered by a set of overlapping squares/cubes. Then, each pixel/voxel is assigned to one of the overlapping patches covering it. As for [41], Shen et al. use a set of selected seeds to generate initial superpixels by performing a lazy random walk. Then, they iteratively update the over-segmentation by optimizing an energy function. Both previously mentioned techniques [41, 47] suffer from a high computational cost. Liu et al. [26] successfully produced superpixels with high boundary adherence by optimizing a new objective function. It combines two terms: an entropy rate of a random walk on a graph and a balancing term. However, the generated superpixels show irregular and non-compact shapes.
A great number of methods adopted a clustering-based approach. They basically rely on iteratively refining an initial clustering until a convergence criterion is fulfilled.
Different techniques have been introduced to generate the initial clustering. The most popular and simple way is to regularly place a set of seeds on the image as adopted in [1, 24, 25, 28, 30, 49]. Seedless approaches were conducted by initially partitioning the image into grid-like superpixels in SEEDS [46] and by adopting a DBSCAN clustering algorithm in [42].
Levinshtein et al. [24] carried out a geometric-flow approach for Turbopixel generation. Given the initial seeds, the boundaries of superpixels evolve iteratively using a level-set representation. The method generates superpixels with regular shapes. However, it achieves a low boundary adherence at high complexity. In [1, 42, 46], low computational cost and good boundary adherence are achieved. The SEEDS technique [46] performs a hill-climbing algorithm to iteratively refine the clustering by exchanging blocks of pixels between neighboring superpixels. In [42], a DBSCAN based algorithm is used to generate superpixels in real-time. Both algorithms (SEEDS and DBSCAN) produce superpixels of irregular shapes. The SLIC algorithm [1] manages to accomplish a compromise between boundary adherence and shape regularity. It is based on a local K-means clustering and has an explicit control on superpixel/voxel compactness. Hence, high quality superpixels/voxels are produced at low memory and computation costs. Inspired by the weighted K-means clustering and Ncuts, Li and Chen [25] introduced a Linear Spectral Clustering based algorithm. It achieves, at a high computational cost, a compact over-segmentation with a high boundary adherence. Machairas et al. [30] introduced Waterpixels, a fast algorithm based on a marker-controlled watershed. The generated superpixels are compact. However, they achieve a low boundary adherence compared to the SLIC algorithm.
All the previously described methods were not designed to adapt the size of superpixel/voxel according to the structures contained in the image. This is of particular interest for images involving objects of different sizes such as medical images. Fewer techniques were proposed to generate structure-sensitive superpixels/voxels. One of the earliest method in this context was introduced by Wang et al. [49]. It combines the Lloyd algorithm and a geodesic distance approximation to generate a structure-sensitive over-segmentation. However, the geodesic distance computation increases the complexity of this method making its extension to the 3D images highly expensive. Lately, Liu et al. [28] introduced the manifold SLIC which extends the SLIC algorithm to generate content-sensitive superpixels. The main idea is based on the generation of a uniform centroidal Voronoi tessellation in a 2D manifold. The latter is created by mapping the initial 5D (position and color) clustering domain of the basic SLIC algorithm. The method shows a high performance in terms of boundary adherence and computation time. We believe that a 3D extension of this method would preserve these properties. However, contrary to the original SLIC algorithm, the additional dimension implies a different formulation of the manifold generation.
In [50], adaptive superpixels are generated on RGB-D images. The proposed technique relies on depth images to generate a density function which is used to produce a non-uniformly distributed samples. Then, a modified SLIC algorithm is applied to create adaptive clusters. The proposed technique shares several similarities with our approach. However, the major difference is that our approach relies uniquely on the gray level information to generate initial sampling.
Despite the significant interest of adaptive supervoxels for medical images analysis, this topic has not been fairly explored. To the best of our knowledge, the few techniques proposed in the literature [3, 7, 53] are limited to generating multi-scale supervoxels. However, adaptive supervoxels are used for offline video processing by Chenliang et al. [54]. In this context, video sequences are considered as a space-time volume. The uniform entropy slice (UES) is used to flatten hierarchies of supervoxels. The proposed method retains the maximum amount of information in the over-segmentation by selecting supervoxels from multiple hierarchical levels. The method shows a good boundary adherence. However, the generated supervoxels exhibit a low compactness. Moreover, the proposed technique requires a high computation time since it starts by generating a supervoxel hierarchy which is afterwards flattened.
In this work, we aim to produce a compact content-sensitive over-segmentation of volumetric images. To achieve this purpose, we use the SLIC algorithm. This choice is motivated by the simplicity, efficiency of this algorithm, as well as the easiness of its 3D extension. Thus, we adopt a clustering based approach with a content-sensitive seed initialization. A PDS is performed using a distance map to image boundaries. We claim that our adaptive over-segmentation efficiently captures the different levels of details in the image while reducing the number of generated supervoxels.
3 Preliminaries
Our AdaSLIC technique relies on two main components: SLIC algorithm [1] and PDS [9]. Therefore, we begin by briefly describing both of them before detailing our algorithm.
3.1 SLIC
The SLIC algorithm [1] uses a local k-means clustering approach to generate supervoxels. Thanks to its good performances, SLIC is widely used in 2D and 3D image processing.
The algorithm starts by regularly placing a set of seeds in the image domain. These seeds are the initial cluster centers that iteratively form the supervoxels. Let Nv be the number of voxels in the image and nsv the desired number of supervoxels. Initial cluster centers are selected on a regular grid with a sampling step \(S=\sqrt [3]{N_{v}/n_{sv}}\) voxels in all three dimensions. The value of S defines the potential size of the supervoxels.
In order to avoid the image edges, the selected seeds are relocated to the lowest gradient position within a neighborhood of 3 × 3 × 3. For each cluster center, its distances to all voxels within a window of 2S × 2S × 2S are computed. Each voxel of the image is then assigned to the cluster corresponding to the closest center. The normalized Euclidean distance between a cluster center ci and a voxel v is defined in (1.3). It combines a spatial distance term ds defined by (1.2) and an intensity difference term dI defined by (1.1).
where I(v), is the gray level attributed to a voxel v.
The distance d(v,ci) uses two normalization constants S and m. As previously mentioned, S accounts for the maximal spatial distance within a cluster, while m balances the influence of intensity similarity with respect to spatial proximity [1]. Small values of m produces supervoxels that adhere more tightly to image boundaries. However, they present irregular shapes and more torturous boundaries.
The SLIC method uses a post-processing step in order to explicitly enforce the connectivity of supervoxels. In this step a connected components algorithm is performed in order to reassign small disjoint segments of supervoxels to the largest neighboring cluster.
3.2 Poisson-disk sampling
Sampling is the process of mapping a continuous function into a set of points. It is a fundamental topic in computer graphics and has been used in different applications such as rendering, stippling, halftoning, mesh generation and texture synthesis [6, 10, 34].
Blue noise sampling has been widely used in the literature because of the visually appealing patterns it produces. It refers to a noise with a minimal low frequency component. Blue noise sampling practically generates a uniform and random distribution of points. These distributions were originally produced to overcome aliasing artifacts of regular sampling, such as Moiré patterns.
Various PDS algorithms have been introduced in order to generate blue noise sampling [4, 12, 13]. Given a uniform domain distribution, a PDS is a set of samples where no pair of samples are closer to each other than a distance r (the Poisson-disk radius). Each sample covers a disk D(r) of radius r and inhibits additional samples in D(r). A maximal Poisson-disk sampling (MPS) is a Poisson-disk sampling where the domain is completely covered by the samples and no more samples could be added. In a non-uniform domain distribution, a PDS is obtained by smoothly varying the samples radii [32].
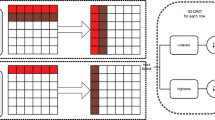
Generally, PDS is performed by sequentially throwing a set of darts in the domain. A dart is accepted as a sample if it fulfills the free disk property. As shown in Fig. 1, a dart of radii ri is rejected if it lies within a distance d < max(ri,rj) from an already accepted sample sj. The uniform sampling is a particular case where ri = rj = r. Dart throwing is performed until a number of consecutive rejections is reached. In order to reduce the complexity of this sampling procedure, different data structures were introduced to track the uncovered regions of the domain (e.g scalloped sectors [12], quadtree [51] and “flat quadtree” [13]).
Disk free check for uniform and variable radii sampling given an initial sample s0. a In a uniform density function, the dart s1 is rejected since d(s0,s1) < r and the dart s2 is accepted since d(s0,s2) > r. b In a non-uniform density function, the dart s1 is rejected since d(s0,s1) < max(r1,r0) , and the dart s2 is accepted since d(s0,s2) > max(r2,r0)
The generalization of the PDS and MPS to higher dimensions has gained a great interest [13]. Yet, few studies treated the generation of an adaptive PDS in volumetric images.
4 AdaSLIC
As a k-mean based algorithm, SLIC starts by selecting a set of seeds representing the initial cluster centers. These seeds are regularly sampled on the image domain regardless of the image content. The first contribution of our work is to make this operation sensitive to the structures in the image by adapting their distribution to the image content. To achieve this purpose, we define a sizing function based on distances to image edges. A PDS is performed with variable radii depending on the sizing function. The generated samples are then used as initial seeds of a modified SLIC. The potential sizes of their corresponding supervoxels depend on their associated radii.
Given an input image, we first apply an enhancement algorithm on the image to reduce the noise and acquisition artifacts that generally affect medical images. The anisotropic filtering proposed by Perona and Malik [37] is used for this purpose. It has the advantage of reducing the image noise while preserving important features and edges.
After filtering, the main steps of our AdaSLIC algorithm are described as follows:
-
1.
Edge Map generation: Canny edge detection algorithm [5] is used. Figure 2c depicts an axial slice of the edge map generated from a knee MRI.
-
2.
Distance Map generation: the Euclidean distance transform to the Edge map is computed.
-
3.
Radius Map computation: A non-uniform distribution function is derived from the Distance map and used to control the sampling density.
-
4.
Adaptive Poisson Disk Sampling.
-
5.
Adaptive Clustering.
These steps are illustrated in Fig. 2 and will be further detailed in the next subsections. We note that the measuring unit of the distance D(v), the radii R(v) and the supervoxel size S is the voxel.
4.1 Distance map (D)
For each voxel v, D(v) denotes the Euclidean distance [19] to the nearest edge voxel. Hence, voxels lying inside large homogeneous region are characterized by high distance values (bright regions in Fig. 2d). However, low values of D are attributed to voxels near the borders and within small structures (dark regions in Fig. 2d).
4.2 Radius map (R)
The radius map is the sizing function used to control the sampling density. It defines the Poisson-disk radius for each voxel. The objective of our adaptive over-segmentation is first to produce dense and small supervoxels in tiny regions and in the vicinity of contours. Second, sparse and large supervoxels are required within big and homogeneous regions. To achieve this purpose, the radius map R(v) of a voxel v is derived from the distance map D(v) as formulated in (2). Distance values D lying in the interval [0,max(D)], are mapped to radii in the interval [0,Rmax].
Rmin and Rmax respectively denote the minimal and the maximal radii. \(D_{R_{min}}\) is the distance value associated to Rmin and max(D) refers to the maximal distance value in D. Rmax could be set as a function of max(D) or for more flexibility could be a user defined parameter.
4.3 Adaptive poisson-disk sampling
Ebeida et al. [13] introduced a multi-level sampling algorithm to generate a uniform MPS algorithm in high dimensions. The sampling process uses a base grid of uniform square cells (of size s0) to store the samples and a “flat quadtree” list to track the active cells (AC) in the grid.
At each level i, the Active Cell List (ACL) contains a list of free, uncovered and equally sized cells (s0/2i). A number of darts are thrown uniformly in the ACL. For each new dart, the disk free property is checked in order to approve its insertion as a new sample. If the dart is accepted, its corresponding cell is removed from the ACL. At the end of each level, the remaining ACs are subdivided and the uncovered subcells are updated for the next level.
Inspired by this approach, we define an adaptive Poisson-disk sampling algorithm. Similar to [13], we use two data structures to track the sampling progress: a grid of uniform cubic cells (of size Rmin) to store the samples and an ACL to track the active cells (AC). However, we adopt a hierarchical approach in the sampling process. We start the algorithm by computing the mean radii of all the grid cells. Then, at each level of the hierarchy, we select the cells to be considered for sampling at this level based on their mean radii. At the first level, cells with low mean radii (located at regions of high density) are selected. Dart throwing is then performed within these cells. At the next levels, higher mean radii are iteratively considered until covering all cells. Expressly, we compute a set of intervals of mean radii \(]R_{l_{i}}, R_{l_{i + 1}}]\). These intervals are used to define the list ACi of eligible ACs for each level i in the hierarchy as shown in (3).
where i = 0,..,nblev.
At a level i, the algorithm proceeds by randomly selecting a cell from ACi. Then similar to Bridson algorithm [4], up to maxTries darts are thrown in the cell. For each dart, if the sample radius is in the interval \(]R_{l_{i}}, R_{l_{i + 1}}]\), we check the disk free property. If the condition is fulfilled, the dart is accepted as a new sample. Otherwise, another dart is thrown. By the end of iterations, the cell become inactive either because a new sample was inserted or because no sample could be inserted. In order to avoid samples on contours (similar to the SLIC algorithm), we reject darts with radii equal to zero. Figure 3 displays the results of three sampling levels (nblev = 3) on a knee MRI.

4.4 Adaptive clustering
Given a set of ns samples defined by their positions and radii {(pi,Ri),i = 1,..,ns}, we perform an adaptive over-segmentation by bringing two modifications to the basic SLIC algorithm. First, we initialize the cluster centers by the set of samples {ci = pi,i = 1,..,ns}. Rather than using a fixed sampling step S for each cluster center ci, we define a variable search window. Its size is 2Si × 2Si × 2Si, where Si is equal to max(floor(Ri),Rmin). In the remainder of this paper, we refer to Si as the size extend of a supervoxel svi. We define Smin as the minimum extend size (Smin = Rmin). Second, we modify the normalized distance between each seed and its neighboring voxels to take into account the variable size extend of the supervoxels. For a cluster center ci with a corresponding size extend Si, the distance to a voxel v is computed as follows:
The adaptive clustering algorithm is summarized in Algorithm 2. Similar to the SLIC algorithm, we add a post-processing step to explicitly enforce the connectivity of supervoxels [1].

5 Experimental results
We implemented our algorithm in C++ and tested it on a PC with an Intel I7-3770 (3.4 GHz) processor and a RAM of 8GB. We visualize the over-segmentation results and the input images using the ITK-Snap software [55].
We carried out our evaluations on two sets of 3D medical images. The first one is a set of 45 knee MR images from the SKI10 dataset [17]. All the images were acquired in the sagittal plane with a pixel spacing of 0.4 × 0.4 mm and a slice distance of 1 mm. They have an average size of 270 × 357 × 106 voxels. The images were segmented interactively by experts at Biomet, Inc. The reader can refer to [17] for further details.
The second set of data is provided by the HVSMR 2016 challenge [35]. It englobes 10 3D CMR images acquired in an axial view by a 1.5T scanner (Phillips Achieva). Manual segmentation of the blood pool and ventricular myocardium was performed by a trained rater and validated by two clinical experts as reported in [35]. The dimension and spacing varies across the images and their average are respectively 124 × 179 × 149 voxels and 0.9 × 0.9 × 0.85 mm.
The ground truth segmentation partially covers the image domain in both datasets. For the knee MRIs, only the femur, the tibia and their surrounding cartilage were labeled (Fig. 4). As for the CMR images, the segmentation covers the blood pool and the ventricular myocardium. Therefore, in our evaluation we distinguish the total number of supervoxels generated in the image (nsv) from the number of supervoxels covered by the ground truth segmentation (nsvSeg).
Ground truth segmentation of a knee MRI. a Axial slice. b Sagittal slice. c Coronal slice. d 3D surface reconstruction generated by ITK-Snap [55]
5.1 Parameters
Our algorithm uses three parameters (Rmin, Rmax and \(D_{R_{min}}\)) to control the number and the sizes of supervoxels. As reported in Figs. 5 and 6, these caracteristics are highly affected by Rmin. However, the impact of Rmax is less significant on the supervoxel density (Fig. 7). This parameter mainly affects the gradation of supervoxel sizes. The parameter \(D_{R_{min}}\) reflects the size of the smallest detail that can be represented by the over-segmentation. For most of our evaluations, Rmax = 0.8 × max(D) and \(D_{R_{min}} = 0.5 \times R_{min}\) unless otherwise noted.
In addition to the above-mentioned parameters, the resulting over-segmentation is affected by the edge detection step. If all the image contours are detected, a dense sampling will be achieved leading to a high number of supervoxels. However, if the edge detection generates sparse contours, distance map will be inaccurate and details can be under-segmented. The algorithm has also two additional parameters: m, the intensity normalization coefficient inherited from the original SLIC algorithm and maxTries, the maximum number of throws during the PDS. In our experiments, m is set to 50 while maxTries is set to 5 in order to achieve a compromise between computational time and efficiency.
5.2 Evaluation
To assess the efficiency of our algorithm, we compare it to the SLIC [29] and to a 3D extension of DBSCAN based algorithm [42]. We run AdaSLIC and SLIC algorithms with the same intensity normalization parameter (m = 50). As for Shen et al. [42] algorithm, we use their suggested parameters and a 6-Neighborhood adjacency in the clustering and merging steps.
Figure 8, illustrates examples of over-segmentations issued by the three algorithms. It is noticeable that our algorithm produces a content-sensitive supervoxels while maintaining a good boundary adherence and a compact supervoxel shape. The DBSCAN and SLIC algorithms generate supervoxels of uniform size, whereas AdaSLIC generates an adaptive over-segmentation. We highlight the presence of small supervoxels within thin structures such as the cartilage and the presence of big supervoxels inside large structures such as the tibia and the femur bones. Moreover, while supervoxels generated by AdaSlic and the SLIC algorithm show regular and compact shapes, DBSCAN algorithm generates supervoxels of irregular and complex shapes.
The AdaSLIC algorithm has the advantage of providing a compressed over-segmentation compared to the state of the art techniques. To give evidence of this property, we perform several over-segmentations using different supervoxel sizes. For the SLIC algorithm, the sampling step S lies in the interval [7,..,25], whereas the minimum size extend for AdaSLIC (Smin = Rmin) lies in the interval [6,..,25]. As for the DBSCAN algorithm, the supervoxel size SDBSCAN lies in the interval [73,..,253]. Figure 9 displays the issued results. We can conclude that our algorithm generates the lowest number of supervoxels, while the SLIC algorithm generates the highest one.
Finally, to quantitatively evaluate the quality of our over-segmentation, we adopt five quality metrics: under-segmentation error (UE), achievable segmentation accuracy (ASA), boundary recall (BR) [52], compactness (C) [40] and contour density (CD) [30]. We also compare the runtime performance of the AdaSLIC to those of the SLIC and DBSCAN algorithms. In the following subsections, we respectively define the metrics and analyze the performance of our technique on both datasets.
5.2.1 Metrics definitions
The UE, ASA and BR metrics are used to measure the boundary adherence of the over-segmentation. On the other hand, the C and CD metrics assess the supervoxel shapes. An over-segmentation should achieve a compromise between boundary adherence and shape regularity and smoothness. These metrics are defined as follows:
-
Under-segmentation Error (UE): This metric was introduced in [24] to measure the percentage of pixels/voxels that leak through the ground-truth segment boundaries. Different variants of this metric were proposed in the literature. In our evaluation, we adopted a definition similar to [1]. Given the ground-truth segmentation {gi,i = 1,..,ng} and an over-segmentation SV = {svi,i = 1,..,nsv}, for a segment gi we identify the corresponding set of overlapping supervoxels as {svj ∈ SV : |svj ∩ gi| > r}. The under-segmentation error is expressed as:
$$ UE=\frac{{\sum\limits_{i = 1}^{n_{g}}}\left[\left( {\sum\limits_{sv_{j} \in SV : \left| sv_{j}\cap g_{i}\right|>r}}\left|sv_{j}\right|\right)-\left|g_{i}\right|\right]}{{\sum\limits_{i = 1}^{n_{g}}}\left|g_{i}\right|} $$(5)|s| is the number of voxels within a segment. In [1], r is chosen as 5% of the supervoxel size.
-
Achievable Segmentation Accuracy (ASA): This metric evaluates the highest achievable accuracy of labeling each supervoxel with the ground truth segment with whom it is overlapping the most. A high ASA means that supervoxels comply well with the objects of the image. For a given over-segmentation, the ASA is formulated as:
$$ ASA=\frac{{\sum\limits_{sv_{j}\in SV}}\{max_{g_{i}}(sv_{j}\cap g_{i})\}}{{\sum}_{i = 1}^{n_{g}}\left|g_{i}\right|} $$(6) -
Boundary recall (BR): This metric measures the fraction of ground truth edges falling within at least t voxels of a supervoxel boundary. In our experiments, similar to [1] we chose t = 2 voxels. A high boundary recall value means that very few true edges were missed.
-
Compactness (C): This metric was first introduced in [40]. It expresses the simplicity of a superpixel shape by comparing its boundary to a circle. To achieve this comparison, the isoperimetric quotient of a superpixel (IQ) is used. It is defined as the ratio between the superpixel area and the area of a circle with the same boundary length. The IQ(s) = 1 for a circle and decreases for irregular shapes. We extend this metric to supervoxels by comparing its surface to a sphere. Let sv be a supervoxel with a volume Vsv expressed in number of voxels, Vsph the volume of a sphere with the same surface area as sv and Asv the surface area of sv. The radius of a sphere with surface area Asv is \(r=\sqrt {\frac {A_{sv}}{4 \pi }}\) and the volume of this sphere is \(V_{sph}=\frac {A_{sv}^{3/2}}{6 \sqrt {\pi }}\). The isoperimetric quotient IQ of sv is then equal to:
$$ IQ(sv)=\frac{V_{sv}}{V_{sph}} =\frac{|sv|}{V_{sph}}=\frac{6\sqrt{\pi}|sv|}{A_{sv}^{3/2}} $$(7)For an over-segmentation, the compactness is computed as:
$$ C=\frac{1}{N_{v}} \sum\limits_{sv\in SV }IQ(sv) \times |sv| $$(8) -
Contour Density (CD): This metric was recently defined in [30] to evaluate the complexity of supervoxels boundaries. It is expressed as the total number of voxels on the supervoxels boundary divided by the total number of pixels in the image.
$$ CD=\frac{\frac{1}{2}\times\left|SB\right|+\left|I_{B}\right|}{N_{v}} $$(9)where |SB| is the number of voxels on the supervoxels boundaries and |IB| is the number of voxels on the image borders.
5.2.2 Quantitative analysis
We run the AdaSLIC, SLIC and the DBSCAN algorithms on all images in both datasets and we report the average metric values in Figs. 10 and 11. The three algorithms produces a low UE and a high ASA values as reported in Fig. 10. Out of the three algorithms, AdaSLIC produced the lowest UE for both dataset (Fig. 10a and b). Moreover, it achieves the highest ASA on the knee dataset. However, The SLIC algorithm has a slightly better ASA on the CMR dataset. As for the DBSCAN, it has the lowest performance in terms of ASA and UE, yet it produces the highest BR metric. This is mainly due to two factors. The first one is the distance metric used in the clustering step which is robust to weak edges. The second reason is related to the non-constrained shape of the supervoxels which is expressed by the lowest compactness values and the highest contour density values as plotted in Fig. 11. We highlight that our algorithm produces the same compactness as the SLIC algorithm while achieving the lowest CD. These measures prove that the generated supervoxels exhibit smooth and regular shapes. When combined with a high boundary adherence, a low CD is a desirable over-segmentation property in applications such as image compression. It reflects that the produced over-segmentation presents an accurate and compressed representation of the image.
We also compare the computation time of the three algorithms for different number of supervoxels. Figure 11d and e show the average running time required for the three algorithms on the knee MRIs and the CMRs respectively. The DBSCAN outperforms the SLIC and AdaSLIC in terms of computation time. As for the our algorithm, it has the highest computation time due to the three additional steps compared to the SLIC algorithm: Edge map generation, Distance map generation and Adaptive Poisson-disk sampling. The edge map is produced using the Canny edge detection with a complexity of O(Nv × log(Nv)). A linear algorithm [19] of complexity O(Nv) computes the Distance map. As for the adaptive Poisson-disk sampling, the algorithm performs at most maxTries × nbAC dart throwing. For each thrown dart, the disk free check and the update of active cells require at most each \({R_{max}^{3}}/{R_{min}^{3}} \) iterations. Hence, the sampling algorithm is O(Nv) and the overall complexity of the AdaSLIC algorithm is O(Nv × log(Nv)). Whereas, the complexity of both DBSCAN and the SLIC algorithms is equal to O(Nv).
We notice that the computation time of the DBSCAN significantly increases as the number of supervoxels decreases. In fact the merging step requires longer time to reach a reduced number of supervoxels. As for our algorithm, Fig. 11d and e illustrate that for very high number of supervoxels the computational time increases almost linearly. This is mainly caused by the Poisson-disk Sampling. A high number of supervoxels is reached when small values of Rmin are selected. In these cases, the PDS deals with a higher number of active cells and hence requires more dart throwing.
6 Discussion
Despite the high compactness and boundary adherence achieved by the AdaSLIC supervoxels, a number of issues have to be addressed in order to improve the performance and the scalability of the algorithm.
The first concern is the computation time which is relatively high compared to both SLIC and DBSCAN algorithms. A straightforward solution is to adopt a parallel implementation of the different algorithm steps. In fact, different parallel implementations of the main steps of the algorithm were introduced such as [15] for the Canny Edge detection algorithm, [48] for the EDT computation, [13] for PDS and [39] for a the SLIC algorithm.
Another issue, which will be considered in future work, is the robustness of the method to noise. In fact, since the Radius Map function relies on the Edge map, it is important to achieve a compromise between detecting the main contours in the image and avoiding false edges. Despite that the Canny Edge detection algorithm is designed to tackle this problem, better results could be achieved by using techniques such as [23].
Finally, the AdaSLIC algorithm does not allow the user to explicitly select the number of supervoxels. It rather takes Rmin as a parameter which instead controls the size of the generated supervoxels. Therefore, the algorithm can be improved to add this option. A possible way to achieve this goal is to include the number of generated supervoxels (nsv) in the computation of the density function.
7 Conclusion
In this paper, we present a method for a content-sensitive supervoxel generation. It is an extension of the SLIC algorithm that produces automatic initial seeds and adaptively sets supervoxel sizes. The seeds are selected based on a density function generated from the image contours. An adaptive PDS algorithm is then performed to generate random samples with variable radii. Finally the SLIC algorithm is applied with a modified distance function adequate to the size of the supervoxels. Our AdaSLIC algorithm achieves a high boundary adherence and a high compactness with a reduced number of supervoxels.
The proposed algorithm shares the advantages of the original SLIC algorithm, yet avoids the regular seeds sampling and the uniform supervoxel sizes. The main drawback of the algorithm is the high computational time compared to the SLIC and DBSCAN algorithms. This problem will be addressed in future work by adopting a GPU parallel implementation.
References
Achanta R, Shaji A, Smith K, Lucchi A, Fua P, Süsstrunk S (2012) SLIC superpixels compared to state-of-the-art superpixel methods. IEEE Trans Pattern Anal Mach Intell 34(11):2274–2282
Andres B, Koethe U, Kroeger T, Helmstaedter M, Briggman KL, Denk W, Hamprecht FA (2012) 3d segmentation of SBFSEM images of neuropil by a graphical model over supervoxel boundaries. Med Image Anal 16(4):796–805
Bi L, Kim J, Kumar A, Wen L, Feng D, Fulham M (2017) Automatic detection and classification of regions of FDG uptake in whole-body PET-CT lymphoma studies. Comput Med Imaging Graph 60(Supplement C):3–10
Bridson R (2007) Fast poisson disk sampling in arbitrary dimensions. In: SIGGRAPH sketches, p 22
Canny J (1986) A computational approach to edge detection. IEEE Trans Pattern Anal Mach Intell 8(6):679–698
Chen Z, Yuan Z, Choi YK, Liu L, Wang W (2012) Variational blue noise sampling. IEEE Trans Vis Comput Graph 18(10):1784–1796
Conze PH, Noblet V, Rousseau F, Heitz F, de Blasi V, Memeo R, Pessaux P (2017) Scale-adaptive supervoxel-based random forests for liver tumor segmentation in dynamic contrast-enhanced CT scans. Int J CARS 12(2):223–233
Conze PH, Tilquin F, Noblet V, Rousseau F, Heitz F, Pessaux P (2017) Hierarchical multi-scale supervoxel matching using random forests for automatic semi-dense abdominal image registration. In: IEEE 14th international symposium on biomedical imaging (ISBI), 2017. IEEE
Cook RL (1986) Stochastic sampling in computer graphics. ACM Trans Graph 5(1):51–72
Corsini M, Cignoni P, Scopigno R (2012) Efficient and flexible sampling with blue noise properties of triangular meshes. IEEE Trans Vis Comput Graph 18 (6):914–924
Dang K, Yuan J, Tiong HY (2013) Voxel labelling in ct images with data-driven contextual features. In: 2013 IEEE international conference on image processing, pp 680–684
Dunbar D, Humphreys G (2006) A spatial data structure for fast poisson-disk sample generation. ACM Trans Graph 25(3):503–508
Ebeida MS, Mitchell SA, Patney A, Davidson AA, Owens JD (2012) A simple algorithm for maximal poisson-disk sampling in high dimensions. Comput Graph Forum 31:785–794
Felzenszwalb PF, Huttenlocher DP (2004) Efficient graph-based image segmentation. Int J Comput Vis 59(2):167–181
Gentsos C, Sotiropoulou CL, Nikolaidis S, Vassiliadis N (2010) Real-time canny edge detection parallel implementation for FPGAs. In: 17th IEEE international conference on electronics, circuits, and systems (ICECS), 2010. IEEE, pp 499–502
Grundmann M, Kwatra V, Han M, Essa I (2010) Efficient hierarchical graph-based video segmentation. In: IEEE conference on computer vision and pattern recognition (CVPR), 2010. IEEE, pp 2141–2148
Heimann T, Morrison BJ, Styner MA, Niethammer M, Warfield S (2010) Segmentation of knee images: a grand challenge. In: Proceedings of MICCAI workshop on medical image analysis for the clinic, pp 207–214
Heinrich MP, Simpson IJ, PapieŻ BW, Brady M, Schnabel JA (2016) Deformable image registration by combining uncertainty estimates from supervoxel belief propagation. Med Image Anal 27:57–71
Hesselink WH, Roerdink JBTM (2008) Euclidean skeletons of digital image and volume data in linear time by the integer medial axis transform. IEEE Trans Pattern Anal Mach Intell 30(12):2204–2217
Irving B, Franklin JM, PapieŻ BW, Anderson EM, Sharma RA, Gleeson FV, Brady M, Schnabel JA (2016) Pieces-of-parts for supervoxel segmentation with global context: Application to DCE-MRI tumour delineation. Med Image Anal 32:69–83
Kanavati F, Misawa K, Fujiwara M, Mori K, Rueckert D, Glocker B (2017) Joint supervoxel classification forest for weakly-supervised organ segmentation. In: International workshop on machine learning in medical imaging. Springer, pp 79–87
Kanavati F, Tong T, Misawa K, Fujiwara M, Mori K, Rueckert D, Glocker B (2017) Supervoxel classification forests for estimating pairwise image correspondences. Pattern Recogn 63:561–569
Konishi S, Yuille AL, Coughlan JM, Zhu SC (2003) Statistical edge detection: learning and evaluating edge cues. IEEE Trans Pattern Anal Mach Intell 25 (1):57–74
Levinshtein A, Stere A, Kutulakos KN, Fleet DJ, Dickinson SJ, Siddiqi K (2009) Turbopixels: fast superpixels using geometric flows. IEEE Trans Pattern Anal Mach Intell 31(12):2290–2297
Li Z, Chen J (2015) Superpixel segmentation using linear spectral clustering. In: 2015 IEEE conference on computer vision and pattern recognition (CVPR), pp 1356–1363
Liu MY, Tuzel O, Ramalingam S, Chellappa R (2011) Entropy rate superpixel segmentation. In: CVPR 2011, pp 2097–2104
Liu Y, Stojadinovic S, Hrycushko B, Wardak Z, Lu W, Yan Y, Jiang SB, Timmerman R, Abdulrahman R, Nedzi L et al (2016) Automatic metastatic brain tumor segmentation for stereotactic radiosurgery applications. Phys Med Biol 61 (24):8440
Liu Y, Yu CC, Yu MJ, He Y (2016) Manifold SLIC: a fast method to compute content-sensitive superpixels. In: 2016 IEEE conference on computer vision and pattern recognition (CVPR), pp 651–659
Lucchi A, Smith K, Achanta R, Knott G, Fua P (2012) Supervoxel-based segmentation of mitochondria in EM image stacks with learned shape features. IEEE Trans Med Imaging 31(2):474–486
Machairas V, Faessel M, Cárdenas-Peña D, Chabardes T, Walter T, Decencière E (2015) Waterpixels. IEEE Trans Image Process 24(11):3707–3716
Mahapatra D, Schüffler PJ, Tielbeek JA, Makanyanga JC, Stoker J, Taylor SA, Vos FM, Buhmann JM (2013) Automatic detection and segmentation of crohn’s disease tissues from abdominal MRI. IEEE Trans Med Imaging 32(12):2332–2347
Mitchell SA, Rand A, Ebeida MS, Bajaj C (2012) Variable radii poisson-disk sampling, extended version. In: Proceedings of the 24th canadian conference on computational geometry, vol 5
Moore AP, Prince SJD, Warrell J, Mohammed U, Jones G (2008) Superpixel lattices. In: 2008 IEEE conference on computer vision and pattern recognition, pp 1–8
Ostromoukhov V (2001) A simple and efficient error-diffusion algorithm. In: Proceedings of the 28th annual conference on computer graphics and interactive techniques, SIGGRAPH ’01. ACM, New York, pp 567–572
Pace DF, Dalca AV, Geva T, Powell AJ, Moghari MH, Golland P (2015) Interactive whole-heart segmentation in congenital heart disease. Springer International Publishing, Cham, pp 80–88
Pei Y, Yi Y, Ma G, Guo Y, Chen G, Xu T, Zha H (2017) Finding dense supervoxel correspondence of cone-beam computed tomography images. In: Machine learning in medical imaging - 8th international workshop, MLMI 2017, held in conjunction with MICCAI 2017, Quebec City, QC, Canada, September 10, 2017, Proceedings, pp 114–122
Perona P, Malik J (1990) Scale-space and edge detection using anisotropic diffusion. IEEE Trans Pattern Anal Mach Intell 12(7):629–639
Ren X, Malik J (2003) Learning a classification model for segmentation. In: Proceedings ninth IEEE international conference on computer vision, vol 1, pp 10–17
Ren CY, Reid I (2011) gSLIC: a real-time implementation of SLIC superpixel segmentation. Department of Engineering Science, University of Oxford, p 6
Schick A, Fischer M, Stiefelhagen R (2012) Measuring and evaluating the compactness of superpixels. In: Proceedings of the 21st international conference on pattern recognition (ICPR2012), pp 930–934
Shen J, Du Y, Wang W, Li X (2014) Lazy random walks for superpixel segmentation. IEEE Trans Image Process 23(4):1451–1462
Shen J, Hao X, Liang Z, Liu Y, Wang W, Shao L (2016) Real-time superpixel segmentation by DBSCAN clustering algorithm. IEEE Trans Image Process 25(12):5933–5942
Shi J, Malik J (2000) Normalized cuts and image segmentation. IEEE Trans Pattern Anal Mach Intell 22(8):888–905
Szmul A, Papiez BW, Bates R, Hallack A, Schnabel JA, Grau V (2016) Graph cuts-based registration revisited: a novel approach for lung image registration using supervoxels and image-guided filtering. In: 2016 IEEE conference on computer vision and pattern recognition workshops (CVPRW), pp 592–599
Tian Z, Liu L, Zhang Z, Xue J, Fei B (2017) A supervoxel-based segmentation method for prostate MR images. Med Phys 44(2):558–569
Van den Bergh M, Boix X, Roig G, de Capitani B, Van Gool L (2012) Seeds: superpixels extracted via energy-driven sampling. In: European conference on computer vision. Springer, pp 13–26
Veksler O, Boykov Y, Mehrani P (2010) Superpixels and supervoxels in an energy optimization framework. In: European conference on computer vision. Springer, pp 211–224
Wang YR, Horng SJ (2004) Parallel algorithms for arbitrary dimensional euclidean distance transforms with applications on arrays with reconfigurable optical buses. IEEE Trans Syst Man Cybern B Cybern 34(1):517–532
Wang P, Zeng G, Gan R, Wang J, Zha H (2013) Structure-sensitive superpixels via geodesic distance. Int J Comput Vis 103(1):1–21
Weikersdorfer D, Gossow D, Beetz M (2012) Depth-adaptive superpixels. In: Proceedings of the 21st international conference on pattern recognition (ICPR2012), pp 2087–2090
White KB, Cline D, Egbert PK (2007) Poisson disk point sets by hierarchical dart throwing. In: 2007 IEEE symposium on interactive ray tracing, pp 129–132
Xu C, Corso JJ (2012) Evaluation of super-voxel methods for early video processing. In: 2012 IEEE conference on computer vision and pattern recognition, pp 1202–1209
Xu C, Corso JJ (2016) Libsvx: a supervoxel library and benchmark for early video processing. Int J Comput Vis 119(3):272–290
Xu C, Whitt S, Corso JJ (2013) Flattening supervoxel hierarchies by the uniform entropy slice. In: Proceedings of the IEEE international conference on computer vision, pp 2240–2247
Yushkevich PA, Piven J, Cody Hazlett H, Gimpel Smith R, Ho S, Gee JC, Gerig G (2006) User-guided 3D active contour segmentation of anatomical structures: significantly improved efficiency and reliability. Neuroimage 31(3):1116–1128
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interests
All the authors certify that they have NO affiliations with or involvement in any organization or entity with any financial interest (such as honoraria; educational grants; participation in speakers’ bureaus; membership, employment, consultancies, stock ownership, or other equity interest; and expert testimony or patent-licensing arrangements), or non-financial interest (such as personal or professional relationships, affiliations, knowledge or beliefs) in the subject matter or materials discussed in this manuscript.
Rights and permissions
About this article
Cite this article
Amami, A., Azouz, Z.B. & Alouane, M.TH. AdaSLIC: adaptive supervoxel generation for volumetric medical images. Multimed Tools Appl 78, 3723–3745 (2019). https://doi.org/10.1007/s11042-017-5563-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11042-017-5563-3