Abstract
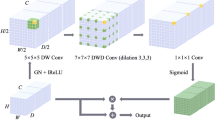
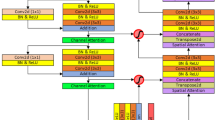
Automatic segmentation of the organ’s tumor and lesion on biomedical imaging is an essential initiative towards clinical study, treatment planning and digital biomedical research. However, precise tumor segmentation on medical imaging is still an open challenge due to the presence of noise in the imaging sequence, the similar tumor pixel intensity with its neighboring tissues, and heterogeneity among human anatomy. Although most of the state-of-the-art algorithms are architecturally dependent on deep convolution networks (DCNs), like 2D and 3D U-Net, they act as a foundation for many biomedical image segmentation. However, 2D DCNs are incompetent to leverage context information from inter-slice completely. At the same time, 3D DCNs can accumulate inter-slice contextual information over the sizeable receptive texture in the organ, but it consumes a considerable amount of GPU memory and burdens with the high execution cost. In order to achieve a promising solution, we proposed a segmentation network called Cascaded Atrous Dual-Attention U-Net. First, our network structure concatenates features from 3D liver segmentation to 2D tumor segmentation for preserving volumetric information as well as enlarging resolution with segmentation accuracy. Second, we embedded dual attention gate in each skip connection layer of the 2D segmentation model, which determines to concentrate on certain discriminative features in order to find tumor segmentation in different organs. Finally, we adopted atrous encoder which extracts wider context features from computed tomography as compared to normal encoder. Furthermore, we tested the proposed method on four different datasets, including liver tumor segmentation benchmark (LiTS), MSD liver, pancreas tumor segmentation and Kidney tumor segmentation (KiTS). Experimental results were compared with the other state-of-the-art segmentation methods. Our proposed approach performs remarkably better than existing methods with around \(4 \sim 6\%\) improvement on each benchmark.







Similar content being viewed by others
References
Albishri AA, Shah SJH, Lee Y (2019) CU-Net: cascaded U-Net model for automated liver and lesion segmentation and summarization. In: 2019 IEEE International conference on bioinformatics and biomedicine (BIBM), IEEE, pp 1416–1423
Alom MZ, Hasan M, Yakopcic C, Taha TM, Asari VK (2018) Recurrent residual convolutional neural network based on u-net (r2u-net) for medical image segmentation. arXiv:1802.06955
Anderson P, He X, Buehler C, Teney D, Johnson M, Gould S, Zhang L (2018) Bottom-up and top-down attention for image captioning and visual question answering. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 6077–6086
Badrinarayanan V, Kendall A, Cipolla R (2017) Segnet: a deep convolutional encoder-decoder architecture for image segmentation. IEEE Trans Pattern Anal Mach Intell 39(12):2481–2495
Bahdanau D, Cho K, Bengio Y (2014) Neural machine translation by jointly learning to align and translate. arXiv:1409.0473
Bilic P, Christ PF, Vorontsov E, Chlebus G, Chen H, Dou Q, Fu CW, Han X, Heng PA, Hesser J, Kadoury S (2019) The liver tumor segmentation benchmark (lits). arXiv:1901.04056
Cai Z, Vasconcelos N (2018) Cascade r-cnn: delving into high quality object detection. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 6154–6162
Cao Y, Xu J, Lin S, Wei F, Hu H (2019) Gcnet: non-local networks meet squeeze-excitation networks and beyond. In: Proceedings of the IEEE international conference on computer vision workshops, pp 0–0
Chartrand G, Cresson T, Chav R, Gotra A, Tang A, DeGuise J (2014) Semi-automated liver CT segmentation using Laplacian meshes. In: 2014 IEEE 11th international symposium on biomedical imaging (ISBI), IEEE, pp 641–644
Chen LC, Papandreou G, Kokkinos I, Murphy K, Yuille AL (2017) Deeplab: semantic image segmentation with deep convolutional nets, atrous convolution, and fully connected crfs. IEEE Trans Pattern Anal Mach Intell 40(4):834–848
Chen LC, Papandreou G, Schroff F, Adam H (2017) Rethinking atrous convolution for semantic image segmentation. arXiv:1706.05587
Chen K, Wang J, Chen LC, Gao H, Xu W, Nevatia R (2015) Abc-cnn: an attention based convolutional neural network for visual question answering. arXiv:1511.05960
Chen L, Zhang H, Xiao J, Nie L, Shao J, Liu W, Chua TS (2017) Sca-cnn: spatial and channel-wise attention in convolutional networks for image captioning. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 5659–5667
Chen LC, Zhu Y, Papandreou G, Schroff F, Adam H (2018) Encoder-decoder with atrous separable convolution for semantic image segmentation. In: Proceedings of the European conference on computer vision (ECCV), pp 801–818
Cheng J, Liu J, Liu L, Pan Y, Wang J (2019) Multi-level glioma segmentation using 3D U-Net combined attention mechanism with atrous convolution. In: 2019 IEEE international conference on bioinformatics and biomedicine (BIBM), IEEE, pp 1031–1036
Christ PF, Elshaer MEA, Ettlinger F, Tatavarty S, Bickel M, Bilic P, Rempfler M, Armbruster M, Hofmann F, D’Anastasi M, Sommer WH (2016) Automatic liver and lesion segmentation in CT using cascaded fully convolutional neural networks and 3D conditional random fields. In: International conference on medical image computing and computer-assisted intervention. Springer, Cham, pp 415–423
Çiçek Ö, Abdulkadir A, Lienkamp SS, Brox T, Ronneberger O (2016) 3D U-Net: learning dense volumetric segmentation from sparse annotation. In: International conference on medical image computing and computer-assisted intervention,. Springer, Cham, pp 424–432
Dolz J, Desrosiers C, Ayed IB (2018) 3D fully convolutional networks for subcortical segmentation in MRI: a large-scale study. NeuroImage 170:456–470
Dou Q, Chen H, Jin Y, Yu L, Qin J, Heng PA (2016) 3D deeply supervised network for automatic liver segmentation from CT volumes. In: International conference on medical image computing and computer-assisted intervention. Springer, Cham, pp 149–157
Fathy ME, Tran QH, Zeeshan Zia M, Vernaza P, Chandraker M (2018) Hierarchical metric learning and matching for 2d and 3d geometric correspondences. In: Proceedings of the european conference on computer vision (ECCV), pp 803–819
Fu J, Liu J, Tian H, Li Y, Bao Y, Fang Z, Lu H (2019) Dual attention network for scene segmentation. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 3146– 3154
Girshick R, Donahue J, Darrell T, Malik J (2014) Rich feature hierarchies for accurate object detection and semantic segmentation. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 580–587
Giusti A, Cireşan DC, Masci J, Gambardella LM, Schmidhuber J (2013) Fast image scanning with deep max-pooling convolutional neural networks. In: 2013 IEEE international conference on image processing, IEEE, pp 4034–4038
Gkika E, Tanadini-Lang S, Kirste S, Holzner PA, Neeff HP, Rischke HC, Reese T, Lohaus F, Duma MN, Dieckmann K, Semrau R (2017) Interobserver variability in target volume delineation of hepatocellular carcinoma. Strahlenther Onkol 193(10):823–830
Hatamizadeh A, Hosseini H, Liu Z, Schwartz SD, Terzopoulos D (2019) Deep dilated convolutional nets for the automatic segmentation of retinal vessels. arXiv:1905.12120
Havaei M, Davy A, Warde-Farley D, Biard A, Courville A, Bengio Y, Pal C, Jodoin PM, Larochelle H (2017) Brain tumor segmentation with deep neural networks. Med Image Anal 35:18–31
He K, Zhang X, Ren S, Sun J (2016) Deep residual learning for image recognition. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 770–778
Heller N, Sathianathen N, Kalapara A, Walczak E, Moore K, Kaluzniak H, Rosenberg J, Blake P, Rengel Z, Oestreich M, Dean J (2019) The kits19 challenge data: 300 kidney tumor cases with clinical context, ct semantic segmentations, and surgical outcomes. arXiv:1904.00445
Holschneider M, Kronland-Martinet R, Morlet J, Tchamitchian P (1990) A real-time algorithm for signal analysis with the help of the wavelet transform. In: Wavelets. Springer, Berlin, pp 286–297
Huang G, Liu Z, Van Der Maaten L, Weinberger KQ (2017) Densely connected convolutional networks. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 4700– 4708
Jiang AW, Liu B, Wang MW (2017) Deep multimodal reinforcement network with contextually guided recurrent attention for image question answering. J Comput Sci Technol 32(4):738–748
Li X, Chen H, Qi X, Dou Q, Fu CW, Heng PA (2018) H-denseUNet: hybrid densely connected UNet for liver and tumor segmentation from CT volumes. IEEE Trans Med Imaging 37(12):2663–2674
Li G, Chen X, Shi F, Zhu W, Tian J, Xiang D (2015) Automatic liver segmentation based on shape constraints and deformable graph cut in CT images. IEEE Trans Image Process 24(12):5315–5329
Li C, Tong Q, Liao X, Si W, Sun Y, Wang Q, Heng PA (2018) Attention based hierarchical aggregation network for 3D left atrial segmentation. In: International workshop on statistical atlases and computational models of the heart. Springer, Cham, pp 255–264
Liao F, Liang M, Li Z, Hu X, Song S (2019) Evaluate the malignancy of pulmonary nodules using the 3-d deep leaky noisy-or network. IEEE Trans Neur Netw Learn Syst 30(11):3484–3495
Lin D, Chen G, Cohen-Or D, Heng PA, Huang H (2017) Cascaded feature network for semantic segmentation of rgb-d images. In: Proceedings of the IEEE international conference on computer vision, pp 1311–1319
Lin TY, Dollár P, Girshick R, He K, Hariharan B, Belongie S (2017) Feature pyramid networks for object detection. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 2117–2125
Lin G, Milan A, Shen C, Reid I (2017) Refinenet: multi-path refinement networks for high-resolution semantic segmentation. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 1925–1934
Linguraru MG, Richbourg WJ, Liu J, Watt JM, Pamulapati V, Wang S, Summers RM (2012) Tumor burden analysis on computed tomography by automated liver and tumor segmentation. IEEE Trans Med Imaging 31(10):1965–1976
Liu YC, Tan DS, Chen JC, Cheng WH, Hua KL (2019) Segmenting hepatic lesions using residual attention U-Net with an adaptive weighted dice loss. In: 2019 IEEE international conference on image processing (ICIP), IEEE, pp 3322–3326
Long J, Shelhamer E, Darrell T (2015) Fully convolutional networks for semantic segmentation. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 3431–3440
Milletari F, Navab N, Ahmadi SA (2016) V-net: fully convolutional neural networks for volumetric medical image segmentation. In: 2016 fourth international conference on 3D vision (3DV), IEEE, pp 565–571
Oktay O, Schlemper J, Folgoc LL, Lee M, Heinrich M, Misawa K, Mori K, McDonagh S, Hammerla NY, Kainz B, Glocker B (2018) Attention u-net: learning where to look for the pancreas. arXiv:1804.03999
Papandreou G, Kokkinos I, Savalle PA (2015) Modeling local and global deformations in deep learning: epitomic convolution, multiple instance learning, and sliding window detection. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 390–399
Ronneberger O, Fischer P, Brox T (2015) U-net: convolutional networks for biomedical image segmentation. In: International conference on medical image computing and computer-assisted intervention. Springer, Cham, pp 234–241
Roth HR, Oda H, Zhou X, Shimizu N, Yang Y, Hayashi Y, Oda M, Fujiwara M, Misawa K, Mori K (2018) An application of cascaded 3D fully convolutional networks for medical image segmentation. Comput Med Imaging Graph 66:90–99
Schlemper J, Oktay O, Schaap M, Heinrich M, Kainz B, Glocker B, Rueckert D (2019) Attention gated networks: learning to leverage salient regions in medical images. Med Image Anal 53:197–207
Sermanet P, Eigen D, Zhang X, Mathieu M, Fergus R, LeCun Y (2013) Overfeat: integrated recognition, localization and detection using convolutional networks. arXiv:1312.6229
Sevilla-Lara L, Sun D, Jampani V, Black MJ (2016) Optical flow with semantic segmentation and localized layers. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 3889–3898
Shen C, Roth HR, Oda H, Oda M, Hayashi Y, Misawa K, Mori K (2018) On the influence of Dice loss function in multi-class organ segmentation of abdominal CT using 3D fully convolutional networks. arXiv:1801.05912
Siegel RL, Miller KD, Jemal A (2019) Cancer statistics, 2019. CA: Cancer J Clinic 69(1):7–34
Simonyan K, Zisserman A (2014) Very deep convolutional networks for large-scale image recognition. arXiv:1409.1556
Simpson AL, Antonelli M, Bakas S, Bilello M, Farahani K, Van Ginneken B, Kopp-Schneider A, Landman BA, Litjens G, Menze B, Ronneberger O (2019) A large annotated medical image dataset for the development and evaluation of segmentation algorithms. arXiv:1902.09063
Snaauw G, Gong D, Maicas G, Van Den Hengel A, Niessen WJ, Verjans J, Carneiro G (2019) End-to-end diagnosis and segmentation learning from cardiac magnetic resonance imaging. In: 2019 IEEE 16th international symposium on biomedical imaging (ISBI 2019), IEEE, pp 802–805
Sudre CH, Li W, Vercauteren T, Ourselin S, Cardoso MJ (2017) Generalised dice overlap as a deep learning loss function for highly unbalanced segmentations. In: Deep learning in medical image analysis and multimodal learning for clinical decision support. Springer, Cham, pp 240–248
Tummala P, Junaidi O, Agarwal B (2011) Imaging of pancreatic cancer: an overview. J Gastrointest Oncol 2(3):168
Van Oostenbrugge TJ, Fütterer JJ, Mulders PF (2018) Diagnostic imaging for solid renal tumors: a pictorial review. Kidney Cancer 2(2):79–93
Velazquez ER, Parmar C, Jermoumi M, Mak RH, Van Baardwijk A, Fennessy FM, Lewis JH, De Ruysscher D, Kikinis R, Lambin P, Aerts HJ (2013) Volumetric CT-based segmentation of NSCLC using 3D-Slicer. Scientif Rep 3:3529
Wang F, Jiang M, Qian C, Yang S, Li C, Zhang H, Wang X, Tang X (2017) Residual attention network for image classification. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 3156–3164
Yang Z, He X, Gao J, Deng L, Smola A (2016) Stacked attention networks for image question answering. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 21–29
Yang Z, Yang D, Dyer C, He X, Smola A, Hovy E (2016) Hierarchical attention networks for document classification. In: Proceedings of the 2016 conference of the North American chapter of the association for computational linguistics: human language technologies, pp 1480–1489
Yu F, Koltun V (2015) Multi-scale context aggregation by dilated convolutions. arXiv:1511.07122
Yu L, Yang X, Chen H, Qin J, Heng PA (2017) Volumetric ConvNets with mixed residual connections for automated prostate segmentation from 3D MR images. In: Thirty-first AAAI conference on artificial intelligence
Zhang Y, Li K, Li K, Zhong B, Fu Y (2019) Residual non-local attention networks for image restoration. arXiv:1903.10082
Zhou XY, Zheng JQ, Li P, Yang GZ (2019) ACNN: a full resolution DCNN for medical image segmentation. arXiv:1901.09203
Acknowledgments
This work was financially supported in part by the center for Cyber-physical System Innovation from The Featured Areas Research Center Program within the framework of the Higher Education Sprout Project by the Ministry of Education (MOE) in Taiwan and Ministry of Science and Technology of Taiwan (MOST109-2218-E-011-010, MOST109-2221-E-011-125-MY3, MOST108-2221-E-011-116), and NTUST-NYMU Joint Research Program (NTUST-NYMU-107-06).
Author information
Authors and Affiliations
Corresponding authors
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Liu, YC., Shahid, M., Sarapugdi, W. et al. Cascaded atrous dual attention U-Net for tumor segmentation. Multimed Tools Appl 80, 30007–30031 (2021). https://doi.org/10.1007/s11042-020-10078-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11042-020-10078-2