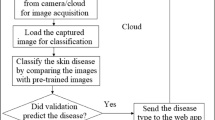
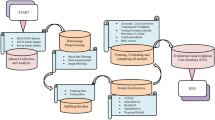
Abstract
A skin lesion is a part of the skin that has abnormal growth on body parts. Early detection of the lesion is necessary, especially malignant melanoma, which is the deadliest form of skin cancer. It can be more readily treated successfully if detected and classified accurately in its early stages. At present, most of the existing skin lesion image classification methods only use deep learning. However, medical domain features are not well integrated into deep learning methods. In this paper, for skin diseases in Asians, a two-phase classification method for skin lesion images is proposed to solve the above problems. First, a classification framework integrated with medical domain knowledge, deep learning, and a refined strategy is proposed. Then, a skin-dependent feature is introduced to efficiently distinguish malignant melanoma. An extension theory-based method is presented to detect the existence of this feature. Finally, a classification method based on deep learning (YoDyCK: YOLOv3 optimized by Dynamic Convolution Kernel) is proposed to classify them into three classes: pigmented nevi, nail matrix nevi and malignant melanomas. We conducted a variety of experiments to evaluate the performance of the proposed method in skin lesion images. Compared with three state-of-the-art methods, our method significantly improves the classification accuracy of skin diseases.








Similar content being viewed by others
References
Alfed N, Khelifi F (2017) Bagged textural and color features for melanoma skin cancer detection in dermoscopic and standard images. Expert Syst Appl 90:101–110
Argenziano G, Fabbrocini G, Carli P, De Giorgi V, Sammarco E, Delfino M (1998) Epiluminescence microscopy for the diagnosis of doubtful melanocytic skin lesions: comparison of the ABCD rule of dermatoscopy and a new 7-point checklist based on pattern analysis. Arch Dermatol 134(12):1563–1570
Arjovsky M, Bottou L (2017) Towards principled methods for training generative adversarial networks. Stat 1050
Arroyo JLG, Zapirain BG, Zorrilla AM (2011) Blue-white veil and dark-red patch of pigment pattern recognition in dermoscopic images using machine-learning techniques. In: 2011 IEEE International Symposium on Signal Processing and Information Technology (ISSPIT), (12):196–201
Aswin RB, Jaleel JA, Salim S (2014) Hybrid genetic algorithm—Artificial neural network classifier for skin cancer detection. In: 2014 International conference on control, instrumentation, Communication and Computational Technologies (ICCICCT), (7):1304–1309.
Celebi ME, Iyatomi H, Stoecker WV, Moss RH, Rabinovitz HS, Argenziano G, Soyer HP (2008) Automatic detection of blue-white veil and related structures in dermoscopy images. Comput Med Imaging Graph 32(8):670–677
Chandy DA, Johnson JS, Selvan SE (2014) Texture feature extraction using gray level statistical matrix for content-based mammogram retrieval. Multimed Tools Appl 72:2011–2024
Chen H, Maharatna K (2020) An automatic R and T peak detection method based on the combination of hierarchical clustering and discrete wavelet transform. IEEE J Biomed Health Inform 24(10):2825–2832
Chen C-C, DaPonte JS, Fox MD (June 1989) Fractal feature analysis and classification in medical imaging. IEEE Trans Med Imaging 8(2):133–142. https://doi.org/10.1109/42.24861
Chen X, Bian X et al (2016) Construction method of uncertain type elementary dependent function in two nested regions. J Inner Mongolia Univ Nationalities 31(3):185–188
Chen X, Bian X et al (2018) Construction method of uncertain type elementary correlation function under three nested regions. J Heilongjiang Univ Sci Technol 28(1):124–128
Chen B et al (2020) Label co-occurrence learning with graph convolutional networks for multi-label chest X-ray image classification. IEEE J Biomed Health Inform 24(8):2292–2302
Choi YH, Tak YS, Rho S, Hwang E (2013) Skin feature extraction and processing model for statistical skin age estimation. Multimed Tools Appl 64:227–247
Codella NC, Gutman D, Celebi ME, Helba B, Marchetti MA, Dusza SW, ... Halpern A (2018) Skin lesion analysis toward melanoma detection: A challenge at the 2017 international symposium on biomedical imaging (isbi), hosted by the international skin imaging collaboration (isic). In: 2018 IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018), (4):168–172
Di Leo G, Fabbrocini G, Paolillo A, Rescigno O, Sommella P (2009) Towards an automatic diagnosis system for skin lesions: estimation of blue-whitish veil and regression structures. In: 2009 6th international multi-conference on systems, Signals and Devices, (3):1–6
Di Leo G, Paolillo A, Sommella P, Fabbrocini G (2010) Automatic diagnosis of melanoma: a software system based on the 7-point check-list. In: 2010 43rd Hawaii international conference on system sciences, (11):1–10
Esteva A, Kuprel B, Novoa RA, Ko J, Swetter SM, Blau HM, Thrun S (2017) Corrigendum: “Dermatologist-level classification of skin cancer with deep neural networks”. Nature (546):686
Feng-Xu G, Wang K-J (2006) Study on extension control strategy of pendulum system. J Harbin Inst Technol 38(7):1146–1149
Florentin S, Victor V(2012) Applications of Extenics to 2D-Space and 3D Space,” The 6th Conference on Software, Knowledge, Information Management and Applications, Chengdu, China, (12):9–11
Frid-Adar M, Diamant I, Klang E, Amitai M, Goldberger J, Greenspan H (2018) GAN-based synthetic medical image augmentation for increased CNN performance in liver lesion classification. Neurocomputing 321:321–331
Ganster H, Pinz P, Rohrer R, Wildling E, Binder M, Kittler H (2001) Automated melanoma recognition. IEEE Trans Med Imaging 20(3):233–239
Gao L, Pan H, Han Q et al (2015) Finding frequent approximate subgraphs in medical image database. IEEE International Conference on Bioinformatics and Biomedicine (BIBM), 1004–1007.
Greenspan H, Van Ginneken B, Summers RM (2016) Guest editorial deep learning in medical imaging: overview and future promise of an exciting new technique. IEEE Trans Med Imaging 35(5):1153–1159
Haralick RM, Shanmugam K, Dinstein I (Nov. 1973) Textural features for image classification. IEEE Trans Syst Man Cybern SMC-3(6):610–621. https://doi.org/10.1109/TSMC.1973.4309314
He B, Zhu X (2005) Hybrid extension and adaptive control. Control Theory Appl 22(2):165–170
Isola P, Zhu JY, Zhou T, Efros AA (2017) Image-to-image translation with conditional adversarial networks. In: Proceedings of the IEEE conference on computer vision and pattern recognition, 1125–1134
Kittler H, Pehamberger H, Wolff K, Binder MJTIO (2002) Diagnostic accuracy of dermoscopy. The Lancet Oncology 3(3):159–165
Krizhevsky A, Sutskever I, Hinton GE (2012) Imagenet classification with deep convolutional neural networks. In: Advances in neural information processing systems (25):1097–1105
Melbin K, Jacob Vetha Raj Y (2021) Integration of modified ABCD features and support vector machine for skin lesion types classification. Multimed Tools Appl 80(6):8909–8929
Mhaske HR, Phalke DA (2013) Melanoma skin cancer detection and classification based on supervised and unsupervised learning. In: 2013 International conference on circuits, Controls and Communications (CCUBE), (12):1–5
Nver HM, Ayan E (2019) Skin Lesion Segmentation in Dermoscopic Images with Combination of YOLO and GrabCut Algorithm. Diagnostics 9(3):72
Pan H, Li P, Li Q et al (2013) Brain CT image similarity retrieval method based on uncertain location graph. IEEE J Biomed Health Inform 18(2):574–584
Pang S et al (2019) A novel YOLOv3-arch model for identifying cholelithiasis and classifying gallstones on CT images. PLoS ONE 14(6):e0217647
Redmon J, Farhadi A (2017) YOLO9000: better, faster, stronger. In: Proceedings of the IEEE conference on computer vision and pattern recognition, 7263–7271
Redmon J, Divvala S, Girshick R, Farhadi A (2016) You only look once: unified, real-time object detection. In: Proceedings of the IEEE conference on computer vision and pattern recognition, 779–788
Ren S, He K, Girshick R, Sun J (2015) Faster r-cnn: towards real-time object detection with region proposal networks. In: Advances in neural information processing systems (28):91–99
Röhrich E, Thali M, Schweitzer W (2012) Skin injury model classification based on shape vector analysis. BMC Med Imaging 12:32. https://doi.org/10.1186/1471-2342-12-32
Roslin SE (2020) Classification of melanoma from Dermoscopic data using machine learning techniques. Multimed Tools Appl 79(5):3713–3728
Roth HR, Lu L, Liu J, Yao J, Seff A, Cherry K, … Summers RM (2015) Improving computer-aided detection using convolutional neural networks and random view aggregation. IEEE Trans Med Imaging 35(5):1170–1181
Seidenari S, Ferrari C, Borsari S, Benati E, Ponti G, Bassoli S, … Pellacani G (2010) Reticular grey-blue areas of regression as a dermoscopic marker of melanoma in situ. Br J Dermatol 163(2):302–309
Setiawan AW, Faisal A (2020) A study on JPEG compression in color retinal image using BT.601 and BT.709 standards: image quality assessment vs. file size. 2020 International Seminar on Application for Technology of Information and Communication (iSemantic), 436–441
Setiawan AW, Faisal A, Resfita N (2020) Effect of image downsizing and color reduction on skin cancer pre-screening. 2020 International Seminar on Intelligent Technology and Its Applications (ISITIA), 148–151
Stoecker WV et al (2011) Detection of granularity in dermoscopy images of malignant melanoma using color and texture features. Comput Med Imaging Graph 35(2):144–147
Tajbakhsh N, Shin JY, Gurudu SR, Hurst RT, Kendall CB, Gotway MB, Liang J (2016) Convolutional neural networks for medical image analysis: full training or fine tuning? IEEE Trans Med Imaging 35(5):1299–1312
Ulhaq A, Khan A, Robinson R (2020) Evaluating faster-RCNN and YOLOv3 for target detection in multi-sensor data. In: Statistics for Data Science and Policy Analysis, 185-193
Vitoria P, Sintes J, Ballester C (2019) Semantic image inpainting through improved Wasserstein generative adversarial networks. 14th International Conference on Computer Vision Theory and Applications
Warsi F, Khanam R, Kamya S, Suárez-Araujo CP (2019) An efficient 3D color-texture feature and neural network technique for melanoma detection. Inform Med Unlocked 17:100176
Wen C (1983) Extension set and non-compatible problems. J Sci Explor 1:83–97
Wen C, Yong S (2006) Extenics, its significance in science and prospects in application. J Harbin Inst Technol 38(7):1079–1086
Yang C(2005) “The Methodology of Extenics”, “Extenics: Its Significance in Science and Prospects in Application,” The 271th Symposium’s Proceedings of Xiangshan Science Conference, 12:35–38
Yang C, Wen C (2007) Extension engineering. Science Press, Beijing
Yang C, Weihua L, Xiaomei L (2011) Recent research Progress in theories and methods for the intelligent disposal of contradictory problems. J Guangdong Univ Technol 28:86–93
YOLOv3 Structure (n.d.), available online on: https://blog.csdn.net/qq_30815237/article/details/91949543. Accessed on 7-10-2020
Yun Y, Gu I (2013) Image Classification by Multi-Class Boosting of Visual and Infrared Fusion with Applications to Object Pose Recognition. Swedish Symposium on Image Analysis (SSBA), (3):14–15
Zarit B, Super B, Quek F (n.d.) Comparison of five color models in skin pixel classification,” Recognition, Analysis, and Tracking of Faces and Gestures in Real-Time Systems, 58–63. https://doi.org/10.1109/RATFG.1999.799224
Zhang X, Zhu X (2019) Vehicle Detection in the Aerial Infrared Images via an Improved Yolov3 Network. 2019 IEEE 4th International Conference on Signal and Image Processing (ICSIP). IEEE
Zhang-China B, Pham-Australia TD (2010) Multiple Features Based Two-stage Hybrid Classifier Ensembles for Subcellular Phenotype Images Classification. Int J Biom Bioinforma 8:554–562
Zhao Yanwei S (2010) Extension Design. Science Press, Beijing
Availability of data and material
All data used in the experiments are from the local hospital. The datasets generated during the current study are available from the corresponding author on reasonable request.
Code availability
The code generated during the current study are available from the corresponding author on reasonable request.
Funding
The paper is supported by the National Natural Science Foundation of China under Grant No.62072135 and No.61672181.
Author information
Authors and Affiliations
Contributions
XB, HP, KZ, PL, LJ and CC conceived of the study. XB, HP and PL performed the collection and label dermoscopy images. XB and LJ carried out the experiment. XB, HP, KZ and CC analyzed the results. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Appendix
Appendix
Abbreviated form | Full form |
|---|---|
YoDyCK | YOLOv3 optimized by Dynamic Convolution Kernel |
BWV | Blue White Veil (a very critical feature which summarized based on expert experience for the diagnosis of malignant melanoma) |
YOLOv3 | the third version of You Only Look Once |
Rights and permissions
About this article
Cite this article
Bian, X., Pan, H., Zhang, K. et al. Skin lesion image classification method based on extension theory and deep learning. Multimed Tools Appl 81, 16389–16409 (2022). https://doi.org/10.1007/s11042-022-12376-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11042-022-12376-3