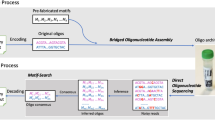
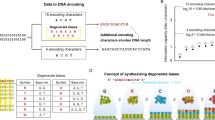
Abstract
A DNA Memory with over 10 million (16.8 M) addresses was achieved. The data embedded into a unique address was correctly extracted through an addressing processes based on nested PCR. The limitation of the scaling-up of the proposed DNA memory is discussed by using a theoretical model based on combinatorial optimization with some experimental restrictions. The results reveal that the size of the address space of the DNA memory presented here may be close to the theoretical limit. The high-capacity DNA memory can be also used in cryptography (steganography) or DNA ink. In decoding process, multiple data with different addresses can be also simultaneously accessed by using the mixture of some address primers.






Similar content being viewed by others
References
Adleman LM (1994) Molecular computation of solutions to combinatorial problems. Science 266:1021–1024
Baum EB (1995) Building an associative memory vastly larger than the brain. Science 268:583–585
Benenson Y, Paz-Elizur T, Adar R, Keinan E, Livneh Z, Shapiro E (2001) Programmable and autonomous computing machine made of biomolecules. Nature 414:430–434
Braich RS, Chelyapov N, Johnson C, Rothemund PWK, Adleman L (2002) Solution of a 20-variable 3-SAT problem on a DNA computer. Science 296:499–502
Chen J, Deaton R, Wang YZ (2005) A DNA-based memory with in vitro learning and associative recall. Nat Comput 4(2):83–101
Clelland CT, Risca V, Bancroft C (1999) Hiding message in DNA microdots. Nature 399:533–544
Deaton R, Murphy RC, Garzon M, Franceschetti DR, Stevens SE Jr (1999) Good encoding for DNA-based solutions to combinatorial problems. In: Landweber LF, Baum EB (eds) DNA based computers II: DIMACS workshop, June 1996, DIMACS series in discrete mathematics and theoretical computer science 44:247–258
Hashiyada M (2004) Development of biometric DNA ink for authentication security. Tohoku J Exp Med 204:109–117
Hashiyada M, Itakura Y, Nagashima T, Nata M, Funayama M (2003) Polymorphism of 17 STRs by multiplex analysis in Japanese population. Forensic Sci Int 133:250–253
Itakura Y, Hashiyada M, Nagashima T, Tsuji S (2002) Proposal on Personal Identifiers Generated from the STR Information of DNA. Int J Information Security 1:149–160
Kameda A, Kashiwamura S, Yamamoto M, Ohuchi A, Hagiya M (2007) Combining randomness and a high-capacity DNA memory. In: Preliminary proceeding of 13th international meeting on DNA Based computers, pp 261–269
Kashiwamura S, Yamamoto M, Kameda A, Shiba T, Ohuchi A (2003) Hierarchical DNA memory based on nested PCR. In: Hagiya M, Ohuchi A (eds) DNA8: 8th International workshop on DNA-based computers, Sapporo, Japan, June 2002. Lecture notes in computer science, vol 2568. Springer, Berlin Heidelberg New York, pp 112–123
Kashiwamura S, Kameda A, Yamamoto M, Ohuchi A (2004) Two-step search for DNA sequence design. IEICE TRANSACTIONS on Fundamentals of Electronics, Commun Computer Sci E87-A(6):1446–1453
Kashiwamura S, Yamamoto M, Kameda A, Shiba T, Ohuchi A (2005a) Potential for enlarging DNA memory: the validity of experimental operations of scaled-up nested primer molecular memory. BioSystems 80:99–112
Kashiwamura S, Yamamoto M, Kameda A, Ohuchi A (2005b) Experimental challenge of scaled-up hierarchical DNA memory expressing a 10,000-address space. In: Preliminary proceeding of 11th international meeting on DNA based computers, p 396
Lipton R (1995) DNA solution of hard combinatorial problems. Science 268:542–545
Lyngso LB, Zuker M, Pedersen CN (1999) Fast evaluation of internal loops in RNA secondary structure prediction. Bioinformatics 15:440–445
McPherson MJ, Hames BD, Taylor GR (1995a) PCR: A practical approach. Oxford University Press
McPherson MJ, Hames BD, Taylor GR (1995b) PCR2: A practical approach. Oxford University Press
Neel A, Garzon MH, Penumatsa P (2004) Improving the quality of semantic retrieval in DNA-based memories with learning. In: KES 2004: 8th Internaltional conference, Wellington, New Zealand, September 20–25 2004, Lecture notes in computer science, vol 3213. Springer, Berlin Heidelberg, New York, pp 18–24
Reif JH, LaBean TH, Pirrung M, Rana VS, Guo B, Kingsford C, Wickham GS (2002) Experimental construction of very large scale dna databases with associative search capability. In: Jonoska N, Seeman NC (eds) DNA 7: 7th International workshop on DNA-based computers, Tampa, FL, USA, June 2001. Lecture notes in computer science, vol 2340. Springer, Berlin, Heidelberg, New York, pp 231–247
SantaLucia J, Allawi HT, Seneviratne PA (1996) Improved nearest-neighbor parameters for predicting DNA duplex stability. Biochemistry 35:3555–3562
Sugimoto N, Nakano S, Yoneyama M, Honda K (1996) Improved thermodynamic parameters and helix initiation factor to predict stability of DNA duplexes. Nucl Acid Res 24:4501–4505
Tanaka F, Kameda A, Yamamoto M, Ohuchi A (2005) Design of nucleic acid sequences for DNA computing based on a thermodynamic approach. Nucl Acid Res 33:903–911
Tulpan DC, Hoos HH, Condon A (2003) Stochastic local search algorithms for DNA word design. In: Hagiya M, Ohuchi A (eds) DNA8: 8th international workshop on DNA-based computers, Sapporo, Japan, June 2002. Lecture notes in computer science, vol 2568. Springer, Berlin, Heidelberg, New York, pp 229–241
Wong PC, Wong KK, Foote H (2003) Organic data memory using the DNA approach. Commun ACM 46(1):95–98
Zuker M, Stiegler P (1981) Optimal computer folding of large RNA sequences using thermodynamics and auxiliary information. Nucl Acid Res 9:133–148
Acknowledgements
We thank M. Hagiya, A. Suyama, A. Kameda and S. Yaegashi for helpful advice and discussions. The work presented in this paper was partially supported by a Grant-in-Aid for Scientific Research on Priority Area No.14085201 and a Grant-in-Aid for Young Scientists (A) No. 17680025, Ministry of Education, Culture, Sports, Science and Technology, Japan.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Yamamoto, M., Kashiwamura, S., Ohuchi, A. et al. Large-scale DNA memory based on the nested PCR. Nat Comput 7, 335–346 (2008). https://doi.org/10.1007/s11047-008-9076-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11047-008-9076-x