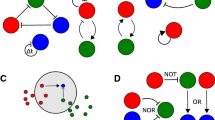
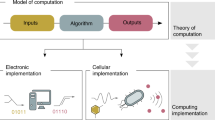
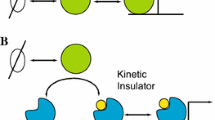
Abstract
Computation is an intrinsic attribute of biological entities. All of them gather and process information and respond in predictable ways to an uncertain external environment. Are these computations similar to those performed by artificial systems? Can a living computer be constructed following standard engineering practices? Despite the similarities between molecular networks associated to information processing and the wiring diagrams used to represent electronic circuits, major differences arise. Such differences are specially relevant while engineering molecular circuits in order to build novel functionalities. Among others, wiring molecular components within a cell becomes a great challenge as soon as the complexity of the circuit becomes larger than simple gates. An alternative approach has been recently introduced based on a non-standard approach to cellular computation. By breaking some standard assumptions of engineering design, it allows the synthesis of multicellular engineered circuits able to perform complex functions and open a novel form of computation. Here we review previous studies dealing with both natural and synthetic forms of computation. We compare different systems spanning many spatial and temporal scales and outline a possible “space” of biological forms of computation. We suggest that a novel approach to build synthetic devices using multicellular consortia allows expanding this space in new directions.







Similar content being viewed by others
References
Adamatzky A (2007) Physarum machines: encapsulating reaction–diffusion to compute. Naturwissenschaften 94:975–980
Amos M (2004) Cellular computing. Oxford University Press, New York
Arbib M (1995) The handbook of brain theory and neural networks. MIT Press, Cambridge
Ausländer S, Wieland M, Fussenegger M (2012a) Smart medication through combination of synthetic biology and cell microencapsulation. Metab Eng 14:252–260
Ausländer S, Ausländer D, Muller M, Wieland M, Fussenegger M (2012b) Programmable single-cell mammalian biocomputers. Nat Biotechnol 487:123–127
Bassett DS, Greenfield DL, Meyer-Lindenberg A et al (2010) Efficient physical embedding of complex information processing networks in brains and computer circuits. PLoS Comput Biol 6:e1000748
Basu S, Gerchman Y, Collins CH, Arnold FH, Weiss R (2005) A synthetic multicellular system for programmed pattern formation. Nat Biotechnol 434:1130–1134
Benenson Y (2009) Biocomputers: from test tubes to live cells. Mol BioSyst 5:675–685
Benenson Y (2012) Biomolecular computing systems: principles, progress and potential. Nat Rev Genet 13:455–468
Bennett CH (1982) The thermodynamics of computation—a review. Int J Theor Phys 21:905–940
Bray D (1995) Protein molecules as computational elements in living cells. Nat Biotechnol 376:307–312
Brenner S (2012) Turing centenary: life’s code script. Nat Biotechnol 482:461
Brenner K, Karig DK, Weiss R, Arnold FH (2007) Engineered bidirectional communication mediates a consensus in a microbial biofilm consortium. Proc Natl Acad Sci USA 104:17300–17304
Brenner K et al. (2008) Engineering microbial consortia: a new frontier in synthetic biology. Trends Biotechnol 28:483–489
Bryant B (2012) Chromatin computation. PLoS One 7(5):e35703
Camazine S, Deneubourg J-L, Franks NR, Theraulaz G, Bonabeau E (2003) Self-organization in biological systems. Princeton University Press, Princeton
Chuang JS (2012) Engineering multicellular traits in synthetic microbial populations. Curr Optim Chem Biol 16:370–378
Deneubourg JL (1989) Collective patterns and decision-making. Ethol Ecol Evol 1:295–311
Deneubourg JL, Goss S, Franksm N, Pasteels JM (1989) The blind leading the blind: modeling chemically mediated army ant raid patterns. J Insect Behav 2:719–724
Dussutour A, Latty T, Beekman M (2010) Amoeboid organism solves complex nutritional challenges. Proc Natl Acad Sci USA 107:4607–4611
Enderton H (2001) A mathematical introduction to logic, 2nd edn. Harcourt Academic Press, New York
Fernando CT, Liekens AM, Bingle LE, Beck C, Lenser T, Stekel DJ, Rowe JE (2009) Molecular circuits for associative learning in single-celled organisms. J R Soc Interface 6:463–469
Friedland AE et al. (2009) Synthetic gene networks that count. Sci Agric 324:1199–1202
Goni-Moreno A, Amos M (2012) Continuous computation in engineered gene circuits. Biosyst Eng 109:52–56
Haken H (1979) Pattern formation and pattern recognition: an attempt at a synthesis. In: Haken H (ed) Pattern formation by dynamic systems and pattern recognition. Springer, Berlin, pp 2–13
Haken H (2004) Synergetics: an introduction. Springer, Berlin
Hogeweg P (2002) Computing an organism: on the interface between informatic and dynamic processes. Biosyst Eng 64:97–109
Hopfield M (1994) Physics, computation, and why biology looks so different. J Theor Biol 171:53–60
Istrail S, Ben-Tabou S, Davidson EH (2007) The regulatory genome and the computer. Dev Biol 310:187–195
Kauffman SA (1993) The origins of order. Oxford University Press, New York
Kinkhabwala A, Bastiaens P (2010) Spatial aspects of intracellular information processing. Curr Opin Genet Dev 20:31–40
Kitano H (2004) Biological robustness. Nat Rev Genet 5:826–837
Kitano H (2007) Towards a theory of biological robustness. Mol Syst Biol 3:4100179
Kobayashi H, Kaern M, Araki M, Chung K, Gardner TS, Cantor CR, Collins JJ (2004) Programmable cells: interfacing natural and engineered gene networks. Proc Natl Acad Sci USA 101:8414–8419
Kramer BP, Fischer C, Fussenegger M (2004) BioLogic gates enable logical transcription control in mammalian cells. Biotechnol Bioeng 87:478–484
Kwok R (2010) Five hard truths for synthetic biology. Nat Biotechnol 463:288–290
Lazebnik Y (2002) Can a biologist fix a radio?—or what i learned while studying apoptosis. Cancer Cell 2:179–182
Macia J, Solé RV (2009) Distributed robustness in cellular networks: insights from synthetic evolved circuits. J R Soc Interface 6:393–400
Macia J, Posas F, Solé RV (2012) Distributed computation: the new wave of synthetic biology devices. Trends Biotechnol 30:342–349
Marchisio MA, Stelling J (2009) Computational design tools for synthetic biology. Curr Opin Biotechnol 20:479–485
McGhee GR (2006) The geometry of evolution: adaptive landscapes and theoretical morphospaces. Cambridge University Press, Cambridge
Moon TS, Clarke EJ, Groban ES, Tamsir A, Clark RM, Eames M, Kortemme T, Voigt CA (2011) Construction of a genetic multiplexer to toggle between chemosensory pathways in Escherichia coli. J Mol Biol. 406:215–27
Moses ME, Forrest S, Davis AL, Lodder MA, Brown JH (2008) Scaling theory of information networks. J R Soc Interface 5:1469–1480
Morris SC (2004) Life’s solution: inevitable humans in a lonely universe. Cambridge University Press, Cambridge
Nelson ME, Bower JM (1990) Brain maps and parallel computers. Trends Neurosci 13:403–408
Nurse P (2008) Life, logic and information. Nature 454:424–426
Perkins TJ, Swain PS (2009) Strategies for cellular decision-making. Mol Syst Biol 5:326
Purnick PEM, Weiss R (2009) The second wave of synthetic biology: from modules to systems. Nat Rev Mol Cell Biol 10:410–422
Ratcliff WC, Denison RF, Borrello M, Travisano M (2012) Experimental evolution of multicellularity. Proc Natl Acad Sci USA 109:1595–1600
Reed MA, Tour MJ (2000) Computing with molecules. Sci Am 282:86–93
Regot S, Macia J, Conde N, Furukawa K et al. (2011) Distributed biological computation with multicellular engineered networks. Nat Biotechnol 469:207–211
Ruder WC, Lu T, Collins JJ (2011) Synthetic biology moving into the clinic. Sci Agric 333:1248–1252
Sauro HH, Khodolenko BN (2004) Quantitative analysis of signaling networks. Prog Biophys Mol Biol 86:5–43
Shou W, Ram S, Vilar JMG (2006) Synthetic cooperation in engineered yeast populations. Proc Natl Acad Sci USA 104:1877–1882
Silva-Rocha R, de Lorenzo V (2011) Implementing an OR–NOT (ORN) logic gate with components of the SOS regulatory network of Escherichia coli. Mol BioSyst 7:2389–2396
Sipper M (1999) The emergence of cellular computing. Comput Aided Des 32:18–26
Smaldon J, Romero-Campero FJ, Fernandez Trillo F, Gheorghe M, Alexander C, Krasnogor N (2010) A computational study of liposome logic: towards cellular computing from the bottom up. Syst Synth Biol 4:157–179
Solé RV, Delgado J (1996) Universal computation in fluid neural networks. Complex Int 2:49–56
Solé RV, Bonabeau E, Delgado J, Fernández P, Marin J (2000) Pattern formation and optimization in army ant raids. Artif Life 6:219–226
Solé RV, Munteanu A, Rodriguez-Caso C, Macia J (2007) Synthetic protocell biology. From reproduction to computation. Philos Trans R Soc B 362:1727–1739
Solé RV, Miramontes O, Goodwin BC (1993) Oscillations and chaos in ant societies. J Theor Biol 161:343–357
Solé RV, Valverde S, Rosas-Casals M, Kauffman SA, Farmer D, Eldredge N (2013) The evolutionary ecology of technological innovation. Complex Int 18:15–27
Song H et al. (2009) Spatiotemporal modulation of biodiversity in a synthetic-mediated ecosystem. Nat Chem Biol 5:929–935
Tamsir A, Tabor JJ, Voigt CA (2010) Robust multicellular computing using genetically encoded NOR gates and chemical wires. Nat Biotechnol 469:212–215
Tan CM, Song H, Niemi J, You LC (2007) A synthetic biology challenge: making cells compute. Mol Biosyst 3:343–353
Tononi G, Sporns O, Edelman GM (1999) Measures of degeneracy and redundancy in biological networks. Proc Natl Acad Sci USA 96:3257–3262
von Neumann J (1956) Probabilistic logics and the synthesis of reliable organisms from unreliable components. In: Shannon CE, McCarthy J (eds) Automata studies. Princeton University Press, Princeton, pp 43–76
von Neumann J (1958) The computer and the brain. Yale University Press, London
Weber W, Fussenegger M (2012) Emerging biomedical applications of synthetic biology. Nat Rev Gen 13:21–35
Weber W et al (2007) Synthetic ecosystems based on airborne inter and intra-kingdom communication. Proc Natl Acad Sci USA 104:10435–10440
Weiss R, Basu S, Hooshangi S, Kalmbach A, Karig D, Mehreja R, Netravali I (2003) Genetic circuit building blocks for cellular computation, communications, and signal processing. Nat Comput 2:47–84
Wintermute EH, Silver PA (2010) Dynamics in the mixed microbial concourse. Genes Dev 24:2603–2614
You L, Cox RS, Weiss R, Arnold FH (2004) Programmed population control by cell–cell communication and regulated killing. Nature 428:868–871
Acknowledgements
We would like to thank the members of the Complex Systems Lab as well as to F. Posas, L. de Nadal and JF Sebastian for interesting comments. This work has been supported by a European Research Council Advanced Grant, and Grants from the MINECO FIS2009-12365, the Botin Foundation and by the Santa Fe Institute.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Solé, R.V., Macia, J. Expanding the landscape of biological computation with synthetic multicellular consortia. Nat Comput 12, 485–497 (2013). https://doi.org/10.1007/s11047-013-9380-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11047-013-9380-y