Abstract
Gene regulatory networks allow single cells to adopt a wide range of different phenotypes in response to changes in environmental conditions. The ecological implications of these cellular computations are poorly understood, and they are largely absent from models of microbial community assembly. Here, we highlight a number of examples where ecological interactions are or may be affected by cellular computations. Our review identifies specific opportunities for integrating cellular decision-making into mathematical models of microbe-microbe interactions and community assembly. We argue that incorporating cellular decision-making into microbial ecology will be critical in order to gain a quantitative understanding of microbial biogeography.
Similar content being viewed by others
1 Introduction
Microorganisms live in dynamical, rapidly changing habitats and are subject to a large number of different environmental stimuli and stresses. Successful and efficient cellular responses to these environmental challenges are crucial to the survival of microbial population. Mounting these phenotypic responses to external stimuli and internal cellular conditions involves a cellular computation, i.e. a selection of one among many alternative cellular phenotypes (i.e. an output) in response to a set of environmental conditions (i.e. inputs). Although they obviously lack the sensory and neural systems that animals have to sense their environment and adopt behavioral responses, single bacterial cells still possess complex biochemical and gene regulatory circuits, which allow cells to sense the state of the environment (the inputs) and switch among multiple well-defined and stable phenotypes in response.
Most microbes live within highly diverse and complex ecological communities. Therefore, the computations carried out by any member of the community may have substantial impact on its ecological partners. For instance, a typical computation amongst soil bacteria involves an input: “nutrient starvation” and the choice of phenotype: “secrete extracellular enzymes that will solubilize nutrients, making them available” as a response to the input. This input–output decision may benefit other species in addition to the producer cells, leading to facilitative ecological interactions (Harrington and Sanchez 2014). Moreover, there is ample evidence that microbial species may directly induce ecologically relevant phenotypic switching in other members of their communities; for instance, members of the Bacillus family induce biofilm formation by closely related species with which they co-occur in nature (Shank et al. 2011).
While clearly very important for microbe-microbe interactions, most mathematical models of microbial community assembly do not explicitly consider the effect of cellular computations. Lotka–Volterra models are phenomenological and typically assume pairwise interaction coefficients that are fixed (Stein et al. 2013; Bashan et al. 2016). Even in cases when more complex relationships are introduced, for instance between the pairwise interaction coefficients and cell density (Sanchez and Gore 2013; Chen et al. 2014), these models typically do not reflect the ability of microbes to sense their environment and dynamically respond to it by altering their behavior. More mechanistic models, such as Consumer-Resource Models (Chesson 1990; Dickens et al. 2016; Posfai et al. 2017) also typically fail to incorporate cellular computations; e.g. when presented with multiple substitutable resources, consumers are typically assumed to utilize them all at the same time. This is in stark contrast with the stereotypical manner in which microbes utilize many substitutable resources, by consuming them one at a time (Aidelberg et al. 2014). This is achieved thanks to gene regulatory networks that sense the concentration of multiple resources, and respond by turning ON only the genes responsible for the metabolism of whichever resource is preferred by the organism (Fig. 1). In other words, microbes perform computations and the precise form of these computations may have important effects on microbial ecological interactions.
Computations allow cells to respond to different inputs that are present at the same time or with a time delay. a Cells often respond to the presence of multiple inputs by activating phenotypic responses to just some of the inputs but not others (Monod 1949, 1966; Venturelli et al. 2015). b When inputs are regularly concatenated in time, microbes can preemptively respond to an input that they have not received yet, poising themselves for when the input finally arrives (Tagkopoulos et al. 2008; Mitchell et al. 2009)
The purpose of this review is to highlight examples where cellular computations may have important ecological effects in microbial communities. We argue that incorporating these computations into ecological theory will benefit our understanding of ecological processes in microbial communities. We will begin by discussing deterministic computations, where all cells in a population respond to the same input stimulus in the same way (Fig. 2a), and discuss how these computations may affect the outcome of ecological competitions and the type of environment created by a microbial population. We will then discuss stochastic computations (Fig. 2b), where gene-regulatory networks allow cells to adopt alternative stable phenotypes in response to the same input (Axelrod et al. 2015), and discuss their relevance for population dispersal and range expansions.
Two forms of response to an input in cellular populations a deterministic, where all cells integrate information about inputs and respond in the same way, or b stochastic, where cells integrate the same input but respond by stochastically adopting two different phenotypes in response (Perkins and Swain 2009). c Often, these are limit scenarios of gene-regulatory networks that may switch from deterministic (monostable) responses to stochastic (bistable) responses depending on the strength of the input. Monostability implies that only one single output phenotype can be stably adopted in response to a given input stimulus. Bistability implies that two possible responses may be stably adopted in response to the same stimulus. In the latter scenario, processes such as temporal fluctuations in the concentrations of regulatory proteins may allow the formation of two subpopulations of cells, one adopting each alternative phenotype. d By way of an example we show the possible phenotypic responses of baker’s yeast (S. cerevisiae) to the availability of galactose in the extracellular medium. The two responses may be to either deterministically turn OFF the galactose utilization genes, or to stochastically turn them ON. The stochastic response regime requires an input strength (Galactose concentration) larger than a threshold value ([galactose] > 0.035% w/v). The deterministic response regime requires the input to be below that threshold. Data from Axelrod et al. (2015)
It is important to note that although stochastic and deterministic computations can be easily distinguished conceptually, in practice they often represent two limit behaviors of the same gene-regulatory network. For instance, metabolic networks in charge of turning ON or OFF metabolic pathways in response to availability of nutrients often exhibit critical transitions between monostable and bi-stable regimes as a function of the strength of the input (Fig. 2c) (Acar et al. 2008; Axelrod et al. 2015). The computation is necessarily deterministic in the monostable regimes, as the network can exist only in one state for a given value of the input. All cells in the population must thus adopt the same phenotype output for the same input. In contrast, bi-stable regimes allow for distinct sub-populations of cells adopting different phenotypes under the exact same input, stochastically switching between them through noise induced fluctuations in gene expression (Fig. 2c). A well-studied example of such a genetic network is the GAL network in yeast (Fig. 2d). At constant glucose concentrations, the network performs a deterministic computation at low galactose concentration, and a stochastic computation at higher galactose levels (Acar et al. 2008; Axelrod et al. 2015).
A final note regarding cellular computations is what may constitute an input. Inputs can be in principle any physical or chemical agents (or combination thereof) that carry out quantitative information about the extra- or intra-cellular environment, and which can be sensed by the cell and propagated through a gene network to elicit a phenotypic response (an output). In the GAL network represented above, the input is the concentration of galactose at constant glucose concentrations. However, recent work has shown that the situation is more complex, and when glucose and galactose are both variable, cells primarily induce the GAL network in response to changes in the ratio between the glucose and galactose concentrations, rather than the absolute amounts of each separate carbon source (Escalante-Chong et al. 2015). Moreover, the input need not even be a concentration. Recent work has shown that cells can also respond to the rate of change of a given chemical stressor, such as salts or ethanol, and even show different responses to the precise functional form of a temporal gradient in a given signal (Young et al. 2013; Li et al. 2017).
2 The ecology of hierarchical carbon source utilization
All cells require carbon to survive, grow, and reproduce. Natural systems generally contain many different types of carbon sources and bacteria have evolved cellular decision making strategies to utilize preferred carbon sources over non-preferred carbohydrates. The decision to utilize one carbon source over another.
In a combination of multiple sources of carbon, bacterial growth is multi-phasic (Fig. 3). Typically one of the carbon sources is exhausted first, and only when it is depleted, bacteria switch phenotypes (expressing the required metabolic machinery) and starts consuming other sources. Metabolic switching in response to resource depletion involves a computation: cells sense their environment (i.e. the concentration of different carbon sources), and respond by activating and inactivating the metabolic pathways that allow them to maximize growth in that environment. The utilization of a preferred carbon source involves complex metabolic mechanisms, often termed carbon catabolite repression (CCR). CCR usually involves the repression of other carbon sources or carbon utilization genes or operons when a preferred carbon source is present.
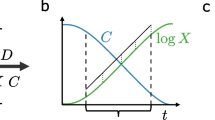
Ecological effects of hierarchical choice among carbon sources. a Unicellular organisms often consume substitutable resources one at a time. In the presence of multiple carbon sources, bacteria will first utilize their preferred source (R1). After R1 is depleted, microbes are faced with an environment that contains two other carbon sources, R2 and R3. In many species, this will lead to a decision over which of the two carbon sources they will consume (e.g. R2). Only when R2 is depleted, the bacteria will eat R3. Hierarchical carbon utilization may lead to coexistence. b Simulation of the model in Eq. 1–4 with parameters f = 2, u = 1, w = 0.001, K = 1010, Jchem = 1, n = 5. This corresponds to a situation where both species S and s uptake both resources with equal rate per molecule regardless of their concentrations, but S converts them to biomass with twice the efficiency than s. c Same as before except for K= 100. In this case, S prefers resource R1 over R2, and will only consume R2 if R1 falls below R1 ~ 100; and s prefers resource R2 over R1, and will only consume R1 if R2 falls below R2 ~ 100
Hierarchical carbon source utilization may produce very different ecological dynamics than the simultaneous utilization that is often assumed in consumer-resource models. For instance, in Fig. 3, we simulate the growth of two species (S, s) on two resources (R1, R2) in a chemostat. We use the following Consumer-Resource Model:
where u is the uptake rate; w is the amount of biomass produced per unit of resource consumed; f is a multiplicative factor that captures the fold-difference in efficiency between species S and species s; Jchem is the flow rate from the reservoir; and the parameter K captures the computations performed by both consumer species: Species S will not consume resource R2 unless its preferred resource R1 has fallen below K. Likewise, species s will not consume resource R1 unless its preferred resource, R2, falls below K. When K is infinite (so R1, R2 << K), then both species consume both resources regardless of their concentrations. In that limit, the most efficient species S outcompetes the less efficient species s (Fig. 3a). However, when K is finite and species are hierarchically choosing between the two carbon sources, both species coexist (Fig. 3b). Of course this result is contingent on the precise type of computations performed by each species. If both species always choose the same type of carbon source over the other, their ability to coexist will be less than if they choose different carbon sources.
Interestingly, different microbial species have indeed different carbon source preferences: even pairs of species with wide metabolic overlap, vary in their carbon source preferences. For instance, the well-studied model organisms E. coli and B. subtilis (Singh et al. 2008; Aidelberg et al. 2014), preferentially utilize glucose over all other sugars and have a detailed carbon hierarchy, preferring simple sugars over more complex carbohydrates (Aidelberg et al. 2014; Beisel and Afroz 2015). This decision is implemented by complex and hierarchical gene-regulatory circuits, controlled by different master regulators in each species (CRP and EII for E. coli; HPr and CcpA for B. subtilis) but leading to very similar hierarchies and even general mechanisms of dietary choice in both: In the presence of the preferred and not preferred carbon sources, the master regulators regulate a genetic regulatory cascade that turns ON the pathways involved in the metabolism of the preferred carbon source, while turning OFF the pathways that would enable the metabolism of the non-preferred carbon sources. While they may be similar in some species, carbon hierarchies are not generally conserved. For instance, Streptococcus thermophilus prefers lactose over glucose (Aidelberg et al. 2014), and Pseudomonadaceae often prefer organic acids and aminoacids over sugars (Rojo 2010). Thus, and although all of these species can utilize a wide gamut of sugars, carboxylic acids and amino acids for growth, and often they are able to utilize overlapping sets of resources, their preferences are different potentially leading to niche separation in practice, in spite of substantial metabolic overlap.
The very process of switching between carbon sources may also have substantial ecological effects. Studies of metabolic phenotype switching at the single cell level have revealed that not all individual cells switch carbon sources at the same time; rather, substantial heterogeneity exists between individuals. For example, in a mixture of the sugars arabinose and xylose, E. coli represses of xylose utilization in favor of arabinose (Desai and Rao 2010; Aidelberg et al. 2014). However, single cell imaging revealed that this population-level repression of xylose utilization hides a more complex set of responses where only some bacteria express just the arabinose utilization operon, while others express the xylose utilization operon, both operons, or neither one (Koirala et al. 2015). Studies across species have suggested this heterogeneity is a result of a tradeoff at the cellular level between growth rate on an initial carbon source and ability to shift to a new carbon source. Venturelli et al. found that on certain glucose-galactose mixtures, Saccharomyces cerevisiae split into two populations- one that switched to galactose early and a second that switched later (Venturelli et al. 2015). However, yeast that express galactose genes were observed to grow around 15% slower than the subpopulation that repressed the genes (Venturelli et al. 2015). Other studies observed the same tradeoff between growth rates on glucose and diauxic lag time when comparing natural isolates of S. cerevisiae (Wang et al. 2015) or experimentally evolved strains (New et al. 2014) rather than subpopulations of genetically identical yeast. A tradeoff has also repeatedly been seen in E. coli between growth rate on glucose and the fraction of cells that switch to utilization of the non-preferred sugar rather than becoming dormant (Robert et al. 2010; Kotte et al. 2014). A tradeoff between growth rate and efficiency in switching to a secondary sugar has thus been observed in a variety of microorganisms. Depending on the circumstances tradeoffs have been observed to contribute to species coexistence in a variety of heterogeneous and homogenous environments (Litchman et al. 2007, 2015; Hall et al. 2010; Bohannan et al. 2002; Beardmore et al. 2011). This further highlights the need to incorporate metabolic decision-making into ecological models of community assembly.
3 Predictive computations
Computations may also be predictive, and produce a response to environmental inputs that have not occurred yet, but which normally and predictably follow other environmental inputs (Fig. 2b). An anticipatory strategy responds to environmental events that occur in a predictable temporal order: an environmental input (priming stimulus) prepares and modifies the cellular response to a future event (triggering stimulus) (Hilker et al. 2016) . Although priming has been well documented in macroorganisms (e.g. Sani et al. 2013) the potential effects of priming on microorganisms has only attracted limited attention until recently (Tagkopoulos et al. 2008; Mitchell et al. 2009; Mitchell and Pilpel 2011; Hilker et al. 2016). An example of predictive computations emerges from the naturally occurring sequence of sugars in the human digestive tract shapes. This shapes an anticipatory biochemical network in Escherichia coli, whose lactose and maltose operon are expressed in an asymmetrical manner: given that the presence of lactose is always followed by the presence of maltose, lactose operons are only induced by lactose, whereas maltose operons are induced both by maltose and, to a lower level, by lactose (Mitchell et al. 2009).
Within a complex multispecies community, interactions between different functional groups may influence whether investment of priming ability pays off over time. Recent theoretical work suggests that, in single species monocultures, the payoff of a predictive response strategy (priming) directly depends on the costs and benefits involved in the habitats: the higher the costs of priming, the higher the benefits have to be (Mitchell and Pilpel 2011). When species interact in silico with a strong competitor, investment in priming may pay off even under high costs (Rillig et al. 2015). These theoretical analyses suggest that ecological interactions could modify the costs and benefits of anticipatory responses found when species grow in monocultures. Rillig et al. (2015) propose three factors that should be considered to analyze the role of priming within a community: the presence, cost, and effectiveness of anticipatory computations. Future experimental work should test these theoretical predictions to give us a clearer picture of the effect of anticipatory computations on community assembly and functioning.
Notably, the temporal changes in environment discussed above are passively experienced by the organisms as part of their external environment. However, microorganisms can also dramatically affect their chemical environment through the uptake and secretion of molecules to the extracellular space (Goldford et al. 2018). Microorganisms may feed on the secondary metabolites produced by other species, leading to cross-feeding. Therefore, microorganisms that are poor competitors in current environmental resources may anticipate the future presence of secondary metabolites produced by other strong competitors (e.g. the fermentation of abundant sugars leads to the production of organic acids). Anticipatory computations may thus compensate for a disadvantage in cellular growth, and may help microorganisms avoid direct and intense competition for current abundant resources, potentially leading to coexistence.
4 Evolution of novel metabolic traits involve evolutionary rewiring of existing computations
Evolution of novel computations causes dramatic changes in the environment and leads to ecological interactions. Microbes readily adapt in order to access new ecological niches. Whilst these adaptations may involve the evolution of new metabolic functions, they often occur exclusively through gene regulatory changes (Toll-Riera et al. 2016). Mutations can affect gene regulation by (i) altering the regulatory functions of constitutively expressed gene; (ii) changing the conditions under which facultatively expressed genes are expressed; (iii) converting facultatively expressed genes into constitutively expressed genes (and vice versa). Moreover, mutations affecting gene regulation can have profound ecological effects (Turkarslan et al. 2011). The idea that computations may strongly affect the environment and lead to ecological interactions is supported by long-term evolution experiments with the bacterium Escherichia coli.
Naturally occurring populations of E. coli are incapable of utilizing citrate under aerobic conditions, a phenotype long viewed as a defining characteristic of the species. However, E. coli is able to utilize citrate in anaerobic conditions if an oxidizable co-substrate is present by expressing citT, a citrate/C4′-dicarboxylate antiporter (Pos et al. 1998). In one of twelve independently evolving populations (ara-3) of a long-term evolution experiment (LTEE), one E. coli lineage evolved the ability to grow on citrate in aerobic conditions (Blount et al. 2008). The critical step in this evolutionary innovation was the tandem duplication of a region including the citT gene that combined a downstream aerobically expressed promoter rnk with an upstream synthase citG (Blount et al. 2012). This hybrid aerobically expressed promoter rnk-citG was positioned upstream of the duplicate citT and conferred weak growth on citrate in aerobic conditions, at the cost of secretion of C4-dicarboxylates such as succinate. Thus, a single change in the regulatory logic of a metabolic promoter, produced massive alterations to the environment, by changing an environment rich in an inert carbon source (citrate), into one rich in readily metabolizable C4-dicarboxylates.
The change in regulatory logic, from facultative expression of citT in anaerobic conditions to the expression of citT in both aerobic and anaerobic conditions, created a new ecological niche to which a second lineage with a Cit- phenotype was able to adapt (Turner et al. 2015). Cit- coexist with the Cit+ for approximately 10,000 generations because Cit- evolves the ability to cross-feed on the C4-dicarboxylates secreted by Cit+.
Whilst the citrate innovation is unique to the ara-3 line, changes affecting acetate metabolism are ubiquitous in the LTEE. Strains isolated from 50,000 generation across all populations on average secreted 50% more acetate (Harcombe et al. 2013). Moreover, mutations in the transcriptional regulators iclR, arcA and arcB repeatedly arose in most populations including ara-3 and are associated with the evolution of improved growth on acetate (Barrick and Lenski 2013). These mutations do not show a cost on glucose implying that the conditions in the LTEE have favored glucose-acetate generalists (Leiby and Marx 2014), probably due to low glucose concentrations (Quandt et al. 2015).
E. coli strains typically exhibit a diauxic shift when growing on a combination of acetate and glucose, firstly consuming glucose and then switching to consume acetate. This switch results in a second lag period (Fig. 3) during which cells produce the enzymes necessary to metabolize the acetate (Monod 1966). When E. coli is evolved in glucose and acetate it repeatedly diversifies into two ecotypes with different lag phases (Friesen et al. 2004). Fast switchers have short lag phase and simultaneously consume glucose and acetate at the cost of reduced glucose growth. In contrast, slow switchers will exclusively grow on glucose and either show a much longer diauxic lag when switching to acetate or in the extreme; will not switch at all due to oxygen limitation (Le Gac et al. 2008).
The evolution of fast switching is associated with changes in gene regulation, though the identity and number of mutations differ across populations. When growing on glucose the fast switcher tends to show upregulated acetate metabolism, as well as upregulation of genes associated with anaerobic respiration (Le Gac et al. 2008). This allows fast switchers to efficiently use both glucose and acetate even when oxygen is limiting. For example, in one well characterized population fast switching is primarily because of a failure to downregulated malate synthase A (aceB), an enzyme in the glyoxylate cycle required for acetate consumptions (Spencer et al. 2007). A loss of function mutation in iclR a negative regulator of the aceBAK operon which contains aceB is responsible for this failed repression. Slow switching strains, whilst phenotypically more similar to the ancestor, also show substantive genetic changes, some of which are parallel and some unique to specific populations (Herron and Doebeli 2013). When growing on glucose the slow switchers tend to show on upregulation of genes associated with acetate excretion and a downregulation of the TCA cycle, allowing for fast glucose consumption when oxygen is abundant (Friesen et al. 2004).
In summary, Mutations in four genes in the ara-3 population of the LTEE experiment have led to gene regulatory changes with diverse ecological effects. Mutations in the promoter regions of two transporters, citT and dctA, are primarily responsible for the evolution of citrate usage and can lead to both the construction of new ecological niches (citT) and their subsequent destruction (dctA). Adaptation to two other carbon sources, succinate and acetate, also involve gene regulatory changes. A dcuS mutations allows the Cit- lineage to consume succinate at the cost of growth on glucose whereas a gltA mutation is involved in the evolution of acetate-glucose generalist and appears to be cost free. Whereas in the LTEE gene regulatory changes have led to the evolution of glucose-acetate generalist, in other E. coli evolution experiments, they are implicated in the emergence of glucose and acetate specialists. This suggests that whilst gene regulatory changes are a general mechanism for niche adaptation, the ecological consequences of this phenomenon are context dependent. In order to occupy new niches evolution can rapidly rewire gene regulatory networks in a manner that leading to both coexistence and competitive exclusion. It remains unclear how this scales to complex community, though it seems likely that rewiring of gene regulatory networks can lead to rapid rewiring of ecological networks.
5 Effect of computations beyond metabolic decision making: computations that affect motility
Motility benefits microbial cells by allowing them to explore their environment, find nutrient patches and to move away from unfavorable conditions. Flagella and pili are the foundation for motility, allowing for swimming, twitching motility, biofilm formation, and adhesion to surfaces. However, cells that opt to express motility machinery expend a large portion of metabolic resources that cannot be allocated to improving reproductive fitness. Costs to motility include susceptibility to ultraviolet radiation and oxidative stress, and reduced biofilm formation and surface attachment. Thus, a tradeoff between growth (exploitation) and dispersal (exploration) is found in microbes (Yawata et al. 2014; Xie and Wu 2014).
One way bacterial cells can manage this tradeoff exploration-exploitation tradeoff is by contingently expressing flagellar production as a function of environmental and growth conditions. For instance, in a recent experiment (Yi and Dean 2016), Escherichia coli cultures were propagated in a cyclical environment, which alternated between selection for exploitation (competitive growth in batch culture) and selection for exploration (capillary selection for chemotaxis). Ancestral cultures grown in this cyclical environment boost their swimming speeds during the period of rapid growth, while speed declines as the carrying capacity is reached. Slowed growth of these early clones provides direct evidence that increasing swimming speed requires diversion of resources away from growth. In successive generations the effect of the tradeoff is eased by a change in behavior as the population evolves toward enhanced, more efficient chemotaxis. These generations reduce their swimming speed during exponential growth, and steadily increase their swimming speed as the carrying capacity is approached, evolving to become better at chemotaxis and overcoming partitioning of energy between chemotaxis and growth (Yi and Dean 2016).
Phenotypically plastic behavior in E. coli was produced by a mutation in FliA, a regulatory protein integral to chemotaxis which regulates expression of over 40 chemotaxis genes. The FliA mutation elevated fitness by increasing growth rate, reducing transcription of flagellar machinery and associated energetic cost, and increasing the motile fraction of the population during chemotactic selection. This single mutation has pleiotropic effects, allowing for plasticity required to adapt to the surrounding dynamic environment (Yi and Dean 2016). E. coli’s ability to switch between enhanced growth and motility contributes to evolution by widening the adaptive landscape, and is a promising step towards understanding how cellular decision-making impacts community assembly.
6 Computations that affect phage-bacterial interactions
As we discuss above, bacteria face changing demands from environments that vary both over space and over time. One of the most severe environmental challenges that bacteria have to adapt to comes from ubiquitous parasitoid bacteriophages (phages). Phages are viruses which infect bacteria, hijack cellular metabolism to replicate, then often lyse the cell, killing their host in the process. Because this places strong selection on bacteria to avoid phage-induced lysis, bacteria have evolved numerous defense mechanisms. For instance, bacteria can mutationally modify the cell surface proteins phages use to attach, change the number of such proteins expressed at a given time, restrict the metabolic processes phage need to attach and replicate, excrete barriers which physically impede phage from contacting the cell surface (eg. exopolysaccharides), or degrade phage nucleic acids once they enter the cell. However, these defense mechanisms often impose an additional cost on the host. Mutations and reduced expression of membrane proteins can inhibit their primary function, reduced metabolism can slow growth and reproduction, production of physical barriers is resource-intensive, and nucleic acid degradation pathways can harm host nucleic acids as well. To ameliorate these costs, while retaining the benefits of defense against phage, bacteria utilize gene-regulatory circuits to express defensive mechanisms only when the risk of phage predation is high.
One way bacteria use decision-making computations against phage is in regulation of surface protein expression. The model bacterium E. coli has receptors for N-acyl-l-homoserine lactone (AHL) quorum-sensing molecules. These AHLs are produced by synthases in many gram-negative bacteria. Despite the fact that E. coli itself cannot produce AHLs, this still provides a mechanism for E. coli to approximate the local density of all gram-negative bacteria. This, then, implies the risk of phage predation: when local cell density is high, phages are likely present. E. coli uses this information to guard against phages: when high concentrations of AHLs are present, wildtype E. coli show reduced adsorption rates of the E. coli λ phage. They achieve this reduced infection rate by downregulating expression of LamB, the surface protein which λ uses as a receptor. This, in turn, increases the survival rate of E. coli exposed to λ approximately three-fold (Høyland-Kroghsbo et al. 2013). However, this quorum sensing driven regulation was not only limited to LamB. Induction of the same regulatory elements by high concentrations of AHL molecules also reduces the adsorption of the broad host range phage chi (χ), which infects E. coli and other enteric bacteria by attaching to the flagella. This is likely associated with the known regulation of flagellar genes by AHL quorum sensing (Høyland-Kroghsbo et al. 2013). Both examples show that the reduced expression of phage target proteins in response to environmental signals of high cell concentrations is an effective defense mechanism against phage infection.
Bacteria also use environmental signals to induce expression of physical barriers to phage infection. One highly effective physical barrier is the production and excretion of aggregative matrices. When cells contact each other or a surface, they adhere, forming a biofilm. This biofilm can then provide physical protection from phages. In one experiment, Tan et al. (2015) showed that the addition of broad-host-range phage strain KVP40 to Vibrio anguillarum strain PF430-3 increased biofilm formation by the bacterium compared to a phage-less control. However, they found that this was not a general response by V. anguillarum to any phage: bacterial strain BA35 showed reduced levels of biofilm formation in response to phage ΦH20. Interestingly, this difference was not the consequence of differences in phage lethality: both phages reduced bacterial densities when grown in liquid culture. Rather, they demonstrated that this effect proceeds from increased aggregation of cells from free-living in liquid culture to attachment as a biofilm. The aggregate matrix formed by these biofilms can then be seen trapping phage particles, effectively preventing them from contacting to susceptible bacterial cells. Thus, the aggregation provides spatial refugia for susceptible cells from the phage (Tan et al. 2015). These results clearly show that bacteria can use environmental signals indicating the presence or likely presence of phage to induce physical protective mechanisms from infection.
An alternate tactic bacteria use to prevent phage infection is to repress all cell metabolism under high-risk conditions. Because bacteriophage need host cell machinery to enter the cell, replicate their genome, and produce new phage proteins, reduced metabolism can prevent or resist an infection. This was clearly demonstrated by Qin et al. with P. aeruginosa and its associated K5 phage (Qin et al. 2017). They experimentally manipulated quorum sensing activation by externally applying quorum sensing molecules or by mutationally modifying the quorum sensing pathway. They found that increased activation decreased phage replication. In particular, phages had a lower burst size (number of phage particles produced per infected host) and lower yields. However, this change was not a result of differential binding. K5 putatively attaches to LPS on the bacterial surface, yet LPS expression did not vary between treatments or over the course of the experiment. More generally, the adsorption rate was not affected by manipulation of the quorum sensing pathway. Instead, under high quorum sensing activation the cells were frequently entering a dormant state. As observed from microscopy, cells that had entered this dormant state were unable to be infected by the phages (Qin et al. 2017). Thus, the bacteria used environmental signals to detect high phage-predation risk conditions, and gained fitness benefits by preferentially entering dormancy only when the risk of phages was high. Modulation of metabolism in response to phages may thus propagate into affecting ecological interactions with species that are not even susceptible to phages, further illustrating the ecological consequences of phage-response computations.
These examples clearly demonstrate that bacteria can, and do, use environmental signals to regular cellular plasticity. Furthermore, they do so in many cases to contend with ubiquitous and severe selection by bacteriophages. Yet, there is likely still much to be discovered in how cellular decision-making is utilized in the global bacteria-phage arms race.
7 Computations that affect host-microbe ecological interactions and pathogenesis
The pathogen Salmonella typhimurium also utilizes a stochastic strategy during infection, forming two subpopulations with differential susceptibility to antibiotics, which allows the genotype to persist in an antibiotic altered environment (Arnoldini et al. 2014). Virulent cells in host tissues with slower growth can form persistent groups of cells while the host is treated with antibiotics (Diard et al. 2014). Despite slow growth, the virulent persister cells benefit the S. typhimurium population as a whole, as they can then recolonize the gut and continue to infect the host (Diard et al. 2014). Due to T3SS-1 expression, the slower growth rate of virulent compared to avirulent cells allows the virulent cells to develop antibiotic tolerance (Arnoldini et al. 2014; Diard et al. 2014). In antibiotic treated host environments, virulent antibiotic-tolerant persisters are selected for (Diard et al. 2014).
When S. typhimurium infects its host, it encounters a plethora of microbes and their defense mechanisms that they must combat in order to establish a population (Ahmer and Gunn 2011). S. typhimurium needs to compete with these bacteria in order to successfully colonize, which effects pathogen growth (Lawley et al. 2008). It has been shown that the host bacteria are important in regulating S. typhimurium infection, disease, and transmissibility (Lawley et al. 2008). A healthy host microbial community restricts S. typhimurium growth, so when the host has a disrupted intestinal microbial community, a large proportion of highly virulent S. typhimurium results (Lawley et al. 2008; Ahmer and Gunn 2011). In order to better compete with the host bacteria, the virulent S. typhimurium subpopulation modifies the environment by creating inflammation in the host (Thiennimitr et al. 2011). Ethanolamine is present in the intestinal lumen, and was shown to support anaerobic growth of S. typhimurium during inflammation (Thiennimitr et al. 2011). In a healthy gut, hydrogen sulfide gets converted to thiosulfate. During inflammation, the thiosulfate is oxidized to tetrahionate, which is a respiratory electron acceptor that enables S. typhimurium to grow anaerobically on ethanolamine (Thiennimitr et al. 2011). Because the host bacteria cannot use ethanolamine to grow, S. typhimurium gains a growth advantage and is thus able to compete (Thiennimitr et al. 2011). The virulent subpopulation of S. typhimurium causes inflammation, which enables the entire population to gain a growth advantage over the host microbiota but using ethanolamine (Thiennimitr et al. 2011). This advantage is especially realized by the avirulent S. typhimurium, which have a faster growth rate than the virulent subpopulation (Thiennimitr et al. 2011; Diard et al. 2013).
8 Discussion
The literature reviewed above strongly suggests that ecological interactions are critically affected by cellular decision-making. Microbial communities are large and highly complex, formed by dozens of species that respond to environmental changes and to each other’s actions by dynamically altering their behavior. Gene-regulatory circuits can be rapidly re-wired by evolution (Taylor et al. 2015), suggesting that ecological interactions may be rapidly rewired too by evolutionary changes in gene-regulatory circuits (Harrington and Sanchez 2014).
In addition, the dependence of ecological interactions on cellular decision-making can also lead to rapid rewiring of ecological networks through non-evolutionary processes. The invasion of a community by a new ecological player (e.g. a bacteriophage) may induce phenotypic responses on the resident species (e.g. changes in their metabolism) which in turn alters their ecological interactions (e.g. cross-feeding of an essential metabolite). This example illustrates how, when ecological interactions can be turned on and off by the expression of a single operon, they may lead to flexible and fluid ecological networks, and the presence of higher order ecological interactions (Harrington and Sanchez 2014; Mayfield and Stouffer 2017; Levine et al. 2017). While the evolution of cellular computations has received substantial attention (Cavaliere and Sanchez 2016), the ecology of cellular computations has received relatively little from the modeling community, and we argue here that there is strong evidence that they can be very important.
As we have discussed above (e.g. Fig. 3), cells modify their environment through their metabolic activity. They can then sense these changes in the environment and compute a response to them too. In cellular populations, which are distributed in space, the metabolic effects in the environment are local, since the metabolites consumed and released must diffuse through space. This may generate subtly different local environments in different parts of a spatially structured population (i.e. a colony), and thus to different sub-populations adopting alternative phenotypes in response to their local environment. Computations may thus be distributed and cells within the same colony may be reacting to each other’s actions, taking as inputs the outputs of other cells. The repercussions of distributed cellular computations, and its relationship with cohesiveness and even multi-cellular behavior remains an open and very exciting question in the field (Regot et al. 2011; Cavaliere and Sanchez 2016). Cellular differentiation within colonies may also be mediated by stochastic computations. An example is the stochastic transition to sporulation in individual cells of a B. subtilis colony, in response to low nutrient conditions. The recent development of computationally efficient individual-cell models (ICMs) (Jang et al. 2012; Gutiérrez et al. 2017) is a very promising tool to interrogate the conditions where cellular communication and division of labor may be beneficial. The ability of ICM packages such as Gro to simulate single-cell behavior within a spatially distributed colony, will be highly beneficial, as they also allows one to explicitly track environmental conditions and feedbacks between single-cell behavior and the environment. This offers unparalleled opportunities to dissect the ecological effects of cellular decision-making at the cellular length-scale where they occur.
The examples discussed above all suggest that computations may give organisms a fitness advantage, as they all carry a benefit. However, it is important to remark that computations also have costs. Sensing external or internal inputs and relaying the information to mount a phenotypic response is typically carried out by two-component systems and other energy consuming signaling pathways (Skerker et al. 2008). Accurately sensing the environment is thermodynamically costly (Mehta and Schwab 2012), and this adds an energetic cost to all cellular computations. Therefore, cellular computations carry a cost in addition to conferring benefits, and this may limit the range and extent of computations, particularly in energy poor environments. Errors in performing computations may also be costly, and when the wrong phenotype is chosen for a particular environmental input, this may reduce the fitness advantage of the computing cell. For instance, yeast cells have been found to misinterpret a novel temporal pattern of a stressor (an oscillatory osmotic shock that yeast has not naturally experienced throughout its evolutionary history) as a gradual increase in the stressor (which is a more naturally occurrence and they have evolved to respond to). Faced with the oscillatory osmotic shock, cells trigger a stronger stress response than needed, which limits their ability to grow (Mitchell et al. 2015). This is an example of how computations can go awry and reduce fitness relative to a non-responding (non-computing) genotype. Less extreme but equally intriguing examples exist of how less accurate computations may be advantageous in certain fluctuating environments (Granados et al. 2017).
The complexity of computations also set a different form of costs. Recent work has found that the number of regulatory genes, which are required to implement computations in single cells, grows faster with genome size than the number of metabolic genes (van Nimwegen 2003; Ranea et al. 2005). This means that for every metabolic gene that is incorporated to the genome, cells must incorporate higher and higher numbers of regulatory genes, such as transcription factors and two component systems. Maintaining and expressing these genes has costs too (van Nimwegen 2003; Ranea et al. 2005). Therefore, the need to regulate the expression of metabolic genes puts an upper-limit to the metabolic repertoire of bacteria, and thus creates the basis for niche differentiation, which has obvious ecological repercussions.
Our goal is not to provide an exhaustive exploration of known ecological effects of microbial computations, and there are many that we did not include for lack of space. Rather, we hope that this review will inspire others to consider how computations may affect ecological interactions and to encourage their inclusion in ecological models.
References
Acar M, Mettetal JT, van Oudenaarden A (2008) Stochastic switching as a survival strategy in fluctuating environments. Nat Genet 40:471–475
Ahmer BMM, Gunn JS (2011) Interaction of Salmonella spp. with the Intestinal Microbiota. Front Microbiol 2:101
Aidelberg G, Towbin BD, Rothschild D et al (2014) Hierarchy of non-glucose sugars in Escherichia coli. BMC Syst Biol 8:133
Arnoldini M, Vizcarra IA, Peña-Miller R et al (2014) Bistable expression of virulence genes in salmonella leads to the formation of an antibiotic-tolerant subpopulation. PLoS Biol 12:e1001928
Axelrod K, Sanchez A, Gore J (2015) Phenotypic states become increasingly sensitive to perturbations near a bifurcation in a synthetic gene network. Elife 4:e07935. https://doi.org/10.7554/eLife.07935
Barrick JE, Lenski RE (2013) Genome dynamics during experimental evolution. Nat Rev Genet 14:827–839
Bashan A, Gibson TE, Friedman J et al (2016) Universality of human microbial dynamics. Nature 534:259–262
Beardmore RE, Gudelj I, Lipson DA, Hurst LD (2011) Metabolic trade-offs and the maintenance of the fittest and the flattest. Nature 472:342–346
Beisel CL, Afroz T (2015) Rethinking the hierarchy of sugar utilization in bacteria. J Bacteriol 198:374–376
Blount ZD, Borland CZ, Lenski RE (2008) Historical contingency and the evolution of a key innovation in an experimental population of Escherichia coli. Proc Natl Acad Sci U S A 105:7899–7906
Blount ZD, Barrick JE, Davidson CJ, Lenski RE (2012) Genomic analysis of a key innovation in an experimental Escherichia coli population. Nature 489:513–518
Bohannan BJM, Kerr B, Jessup CM et al (2002) Trade-offs and coexistence in microbial microcosms. Antonie Van Leeuwenhoek 81:107–115
Cavaliere M, Sanchez A (2016) The evolutionary resilience of distributed cellular computing. In: Membrane computing. Springer, Cham, pp 3–15
Chen A, Sanchez A, Dai L, Gore J (2014) Dynamics of a producer-freeloader ecosystem on the brink of collapse. Nat Commun 5:3713
Chesson P (1990) MacArthur’s consumer-resource model. Theor Popul Biol 37:26–38
Desai TA, Rao CV (2010) Regulation of arabinose and xylose metabolism in Escherichia coli. Appl Environ Microbiol 76:1524–1532
Diard M, Garcia V, Maier L et al (2013) Stabilization of cooperative virulence by the expression of an avirulent phenotype. Nature 494:353–356
Diard M, Sellin ME, Dolowschiak T et al (2014) Antibiotic treatment selects for cooperative virulence of Salmonella typhimurium. Curr Biol 24:2000–2005
Dickens B, Fisher CK, Mehta P (2016) Analytically tractable model for community ecology with many species. Phys Rev E 94:022423
Escalante-Chong R, Savir Y, Carroll SM et al (2015) Galactose metabolic genes in yeast respond to a ratio of galactose and glucose. Proc Natl Acad Sci U S A 112:1636–1641
Friesen ML, Saxer G, Travisano M, Doebeli M (2004) Experimental evidence for sympatric ecological diversification due to frequency-dependent competition in Escherichia coli. Evolution 58:245–260
Goldford JE, Lu N, Bajic D et al (2018) Emergent simplicity in microbial community assembly. Science 361:469–474
Granados AA, Crane MM, Montano-Gutierrez LF et al (2017) Distributing tasks via multiple input pathways increases cellular survival in stress. Elife 6:e21415. https://doi.org/10.7554/eLife.21415
Gutiérrez M, Gregorio-Godoy P, Pérez Del Pulgar G et al (2017) A new improved and extended version of the multicell bacterial simulator gro. ACS Synth Biol 6:1496–1508
Hall EK, Singer GA, Kainz MJ, Lennon JT (2010) Evidence for a temperature acclimation mechanism in bacteria: an empirical test of a membrane-mediated trade-off. Funct Ecol 24:898–908
Harcombe WR, Delaney NF, Leiby N et al (2013) The ability of flux balance analysis to predict evolution of central metabolism scales with the initial distance to the optimum. PLoS Comput Biol 9:e1003091
Harrington KI, Sanchez A (2014) Eco-evolutionary dynamics of complex social strategies in microbial communities. Commun Integr Biol 7:e28230
Herron MD, Doebeli M (2013) Parallel evolutionary dynamics of adaptive diversification in Escherichia coli. PLoS Biol 11:e1001490
Hilker M, Schwachtje J, Baier M et al (2016) Priming and memory of stress responses in organisms lacking a nervous system. Biol Rev Camb Philos Soc 91:1118–1133
Høyland-Kroghsbo NM, Maerkedahl RB, Svenningsen SL (2013) A quorum-sensing-induced bacteriophage defense mechanism. MBio 4:e00362–12
Jang SS, Oishi KT, Egbert RG, Klavins E (2012) Specification and simulation of synthetic multicelled behaviors. ACS Synth Biol 1:365–374
Koirala S, Wang X, Rao CV (2015) Reciprocal regulation of l-arabinose and d-xylose metabolism in Escherichia coli. J Bacteriol 198:386–393
Kotte O, Volkmer B, Radzikowski JL, Heinemann M (2014) Phenotypic bistability in Escherichia coli’s central carbon metabolism. Mol Syst Biol 10:736
Lawley TD, Bouley DM, Hoy YE et al (2008) Host transmission of Salmonella enterica serovar Typhimurium is controlled by virulence factors and indigenous intestinal microbiota. Infect Immun 76:403–416
Le Gac M, Brazas MD, Bertrand M et al (2008) Metabolic changes associated with adaptive diversification in Escherichia coli. Genetics 178:1049–1060
Leiby N, Marx CJ (2014) Metabolic erosion primarily through mutation accumulation, and not tradeoffs, drives limited evolution of substrate specificity in Escherichia coli. PLoS Biol 12:e1001789
Levine JM, Bascompte J, Adler PB, Allesina S (2017) Beyond pairwise mechanisms of species coexistence in complex communities. Nature 546:56–64
Li G, Kesler BK, Thiemicke A et al (2017) Linearly changing stress environment causes cellular growth phenotype. bioRxiv 155267
Litchman E, Klausmeier CA, Schofield OM, Falkowski PG (2007) The role of functional traits and trade-offs in structuring phytoplankton communities: scaling from cellular to ecosystem level. Ecol Lett 10:1170–1181
Litchman E, Edwards KF, Klausmeier CA (2015) Microbial resource utilization traits and trade-offs: implications for community structure, functioning, and biogeochemical impacts at present and in the future. Front Microbiol 6:254
Mayfield MM, Stouffer DB (2017) Higher-order interactions capture unexplained complexity in diverse communities. Nat Ecol Evol 1:62
Mehta P, Schwab DJ (2012) Energetic costs of cellular computation. Proc Natl Acad Sci U S A 109:17978–17982
Mitchell A, Pilpel Y (2011) A mathematical model for adaptive prediction of environmental changes by microorganisms. Proc Natl Acad Sci U S A 108:7271–7276
Mitchell A, Romano GH, Groisman B et al (2009) Adaptive prediction of environmental changes by microorganisms. Nature 460:220–224
Mitchell A, Wei P, Lim WA (2015) Oscillatory stress stimulation uncovers an Achilles’ heel of the yeast MAPK signaling network. Science 350:1379–1383
Monod J (1949) The growth of bacterial cultures. Annu Rev Microbiol 3:371–394
Monod J (1966) From enzymatic adaptation to allosteric transitions. Science 154:475–483
New AM, Cerulus B, Govers SK et al (2014) Different levels of catabolite repression optimize growth in stable and variable environments. PLoS Biol 12:e1001764
Perkins TJ, Swain PS (2009) Strategies for cellular decision-making. Mol Syst Biol 5:326
Pos KM, Dimroth P, Bott M (1998) The Escherichia coli citrate carrier CitT: a member of a novel eubacterial transporter family related to the 2-oxoglutarate/malate translocator from spinach chloroplasts. J Bacteriol 180:4160–4165
Posfai A, Taillefumier T, Wingreen NS (2017) Metabolic trade-offs promote diversity in a model ecosystem. Phys Rev Lett 118:028103
Qin X, Sun Q, Yang B et al (2017) Quorum sensing influences phage infection efficiency via affecting cell population and physiological state. J Basic Microbiol 57:162–170
Quandt EM, Gollihar J, Blount ZD et al (2015) Fine-tuning citrate synthase flux potentiates and refines metabolic innovation in the Lenski evolution experiment. Elife 4:e09696. https://doi.org/10.7554/eLife.09696
Ranea JAG, Grant A, Thornton JM, Orengo CA (2005) Microeconomic principles explain an optimal genome size in bacteria. Trends Genet 21:21–25
Regot S, Macia J, Conde N et al (2011) Distributed biological computation with multicellular engineered networks. Nature 469:207–211
Rillig MC, Rolff J, Tietjen B et al (2015) Community priming—effects of sequential stressors on microbial assemblages. FEMS Microbiol Ecol. https://doi.org/10.1093/femsec/fiv040
Robert L, Paul G, Chen Y et al (2010) Pre-dispositions and epigenetic inheritance in the Escherichia coli lactose operon bistable switch. Mol Syst Biol 6:357
Rojo F (2010) Carbon catabolite repression in Pseudomonas: optimizing metabolic versatility and interactions with the environment. FEMS Microbiol Rev 34:658–684
Sanchez A, Gore J (2013) Feedback between population and evolutionary dynamics determines the fate of social microbial populations. PLoS Biol 11:e1001547
Sani E, Herzyk P, Perrella G et al (2013) Hyperosmotic priming of Arabidopsis seedlings establishes a long-term somatic memory accompanied by specific changes of the epigenome. Genome Biol 14:R59
Shank EA, Klepac-Ceraj V, Collado-Torres L et al (2011) Interspecies interactions that result in Bacillus subtilis forming biofilms are mediated mainly by members of its own genus. Proc Natl Acad Sci U S A 108:E1236–43
Singh KD, Schmalisch MH, Stülke J, Görke B (2008) Carbon catabolite repression in Bacillus subtilis: quantitative analysis of repression exerted by different carbon sources. J Bacteriol 190:7275–7284
Skerker JM, Perchuk BS, Siryaporn A et al (2008) Rewiring the specificity of two-component signal transduction systems. Cell 133:1043–1054
Spencer CC, Bertrand M, Travisano M, Doebeli M (2007) Adaptive diversification in genes that regulate resource use in Escherichia coli. PLoS Genet 3:e15
Stein RR, Bucci V, Toussaint NC et al (2013) Ecological modeling from time-series inference: insight into dynamics and stability of intestinal microbiota. PLoS Comput Biol 9:e1003388
Tagkopoulos I, Liu Y-C, Tavazoie S (2008) Predictive behavior within microbial genetic networks. Science 320:1313–1317
Tan D, Dahl A, Middelboe M (2015) Vibriophages differentially influence biofilm formation by Vibrio anguillarum strains. Appl Environ Microbiol 81:4489–4497
Taylor TB, Mulley G, Dills AH et al (2015) Evolution. evolutionary resurrection of flagellar motility via rewiring of the nitrogen regulation system. Science 347:1014–1017
Thiennimitr P, Winter SE, Winter MG et al (2011) Intestinal inflammation allows Salmonella to use ethanolamine to compete with the microbiota. Proc Natl Acad Sci U S A 108:17480–17485
Toll-Riera M, San Millan A, Wagner A, MacLean RC (2016) The genomic basis of evolutionary innovation in Pseudomonas aeruginosa. PLoS Genet 12:e1006005
Turkarslan S, Reiss DJ, Gibbins G et al (2011) Niche adaptation by expansion and reprogramming of general transcription factors. Mol Syst Biol 7:554
Turner CB, Blount ZD, Mitchell DH, Lenski RE (2015) Evolution and coexistence in response to a key innovation in a long-term evolution experiment with Escherichia coli. bioRxiv 020958
van Nimwegen E (2003) Scaling laws in the functional content of genomes. Trends Genet 19:479–484
Venturelli OS, Zuleta I, Murray RM, El-Samad H (2015) Population diversification in a yeast metabolic program promotes anticipation of environmental shifts. PLoS Biol 13:e1002042
Wang J, Atolia E, Hua B et al (2015) Natural variation in preparation for nutrient depletion reveals a cost-benefit tradeoff. PLoS Biol 13:e1002041
Xie L, Wu X-L (2014) Bacterial motility patterns reveal importance of exploitation over exploration in marine microhabitats. Part I: theory. Biophys J 107:1712–1720
Yawata Y, Cordero OX, Menolascina F et al (2014) Competition-dispersal tradeoff ecologically differentiates recently speciated marine bacterioplankton populations. Proc Natl Acad Sci U S A 111:5622–5627
Yi X, Dean AM (2016) Phenotypic plasticity as an adaptation to a functional trade-off. Elife 5:e19307. https://doi.org/10.7554/eLife.19307
Young JW, Locke JCW, Elowitz MB (2013) Rate of environmental change determines stress response specificity. Proc Natl Acad Sci U S A 110:4140–4145
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Baskerville, M., Biro, A., Blazanin, M. et al. Ecological effects of cellular computing in microbial populations. Nat Comput 17, 811–822 (2018). https://doi.org/10.1007/s11047-018-9708-8
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11047-018-9708-8