Abstract
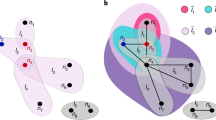
This study provided a game model to simulate the evolution of coauthorship networks, which is a geometric hypergraph built on a circle. A fraction of nodes are randomly selected to attach an arc to express their reputation. The cooperation condition of a new node and existing nodes depends on their distance and the existing nodes’ reputation. The condition gives an expression of kin selection and network reciprocity, two typical mechanisms of cooperation. The size of a node’s reputation is expressed by the length of its arc, which is defined by a function of time and hyperdegree. The function describes the heterogeneity in the size of reputation on nodes and that in the fading speed of reputation on hyperdegrees. The model reveals that the heterogeneities can reproduce the dichotomy of node clustering and degree assortativity, as well as the trichotomy of degree and hyperdegree distributions: generalized Poisson, power-law, and exponential cutoff.





Similar content being viewed by others
References
Barabási, A. L., & Albert, R. (1999). Emergence of scaling in random networks. Science, 286(5439), 509–512.
Barabási, A. L., Jeong, H., Néda, Z., Ravasz, E., Schubert, A., & Vicsek, T. (2002). Evolution of the social network of scientific collaborations. Physica A, 311(3–4), 590–614.
Börner, K., Maru, J. T., & Goldstone, R. L. (2004). The simultaneous evolution of author and paper networks. Proceedings of the National Academy of Sciences of the United States of America, 101(suppl 1), 5266–5273.
Catanzaro, M., Caldarelli, G., & Pietronero, L. (2004). Assortative model for social networks. Physical Review E, 70, 037101.
Clauset, A., Shalizi, C. R., & Newman, M. E. J. (2009). Power-law distributions in emprical data. SIAM Review, 51, 661–703.
Consul, P. C., & Jain, G. C. (1973). A generalization of the Poisson distribution. Technometrics, 15(4), 791–799.
Ductor, L. (2015). Does co-authorship lead to higher academic productivity? Oxford Bulletin of Economics and Statistics, 77(3), 385–407.
Ferligoj, A., Kronegger, L., Mali, F., Snijders, T. A., & Doreian, P. (2015). Scientific collaboration dynamics in a national scientific system. Scientometrics, 104(3), 985–1012.
Glänzel, W. (2014). Analysis of co-authorship patterns at the individual level. Transinformacao, 26, 229–238.
Glänzel, W., & Schubert, A. (2004). Analysing scientific networks through co-authorship. Handbook of quantitative science and technology research, Vol. 11, pp. 257–276.
Kim, J., & Diesner, J. (2016). Distortive effects of initial-based name disambiguation on measurements of large-scale coauthorship networks. Journal of the Association for Information Science and Technology, 67(6), 1446–1461.
Krioukov, D., Kitsak, M., Sinkovits, R. S., Rideout, D., Meyer, D., & Boguñá, M. (2012). Network cosmology. Scientific Reports, 2, 793.
Mali, F., Kronegger, L., Doreian, P., & Ferligoj, A. (2012). Dynamic scientific coauthorship networks (2012) In A. Scharnhorst, K. Börner & P. V. D. Besselaar (Eds.), Models of science dynamics. Springer, pp. 195–232.
Milojević, S. (2010). Modes of collaboration in modern science-beyond power laws and preferential attachment. Journal of the Association for Information Science and Technology, 61(7), 1410–1423.
Moody, J. (2004). The strucutre of a social science collaboration network: Disciplinery cohesion form 1963 to 1999. American Sociological Review, 69(2), 213–238.
Newman, M. (2001a). Scientific collaboration networks. I. Network construction and fundamental results. Physical Review E, 64, 016131.
Newman, M. (2001b). Scientific collaboration networks. II. Shortest paths, weighted networks, and centrality. Physical Review E, 64, 016132.
Newman, M. (2001c). The structure of scientific collaboration networks. Proceedings of the National Academy of Sciences of the United States of America, 98, 404–409.
Newman, M. (2001d). Clustering and preferential attachment in growing networks. Physical Review E, 64(2), 025102.
Newman, M. (2002). Assortative mixing in networks. Physical Review Letters, 89, 208701.
Newman, M. (2004). Coauthorship networks and patterns of scientific collaboration. Proceedings of the National Academy of Sciences of the United States of America, 101, 5200–5205.
Nowak, M. A. (2006). Five rules for the evolution of cooperation. Science, 314(5805), 1560–3.
Papadopoulos, F., Kitsak, M., Serrano, M. A., Boguñá, M., & Krioukov, D. (2012). Popularity versus similarity in growing networks. Nature, 489, 537–540.
Penrose, M. (2003). Random geometric graphs, Oxford studies in probability.
Perc, C. (2010). Growth and structure of Slovenia’s scientific collaboration network. Journal of Informetrics, 4, 475–482.
Perc, M. (2014). The Matthew effect in empirical data. Journal of the Royal Society, Interface, 11, 20140378.
Perc, M., & Szolnoki, A. (2008). Social diversity and promotion of cooperation in the spatial prisoner’s dilemma game. Physical Review E, 77(1), 011904.
Perc, M., & Szolnoki, A. (2010). Coevolutionary games-a mini review. BioSystems, 99(2), 109–125.
Ponomariov, B., & Boardman, C. (2016). What is co-authorship? Scientometrics, 109(3), 1939–1963.
Qi, M., Zeng, A., Li, M., Fan, Y., & Di, Z. (2017). Standing on the shoulders of giants: The effect of outstanding scientists on young collaborators’ careers. Scientometrics, 111(3), 1839–1850.
Santos, F. C., & Pacheco, J. M. (2005). Scale-free networks provide a unifying framework for the emergence of cooperation. Physical Review Letters, 95(9), 098104.
Sarigöl, E., Pfitzner, R., Scholtes, I., Garas, A., & Schweitzer, F. (2014). Predicting scientific success based on coauthorship networks. EPJ Data Science, 3(1), 1–16.
Tomassini, M., & Luthi, L. (2007). Empirical analysis of the evolution of a scientific collaboration network. Physica A, 385(2), 750–764.
Wagner, C. S., & Leydesdorff, L. (2005). Network structure, self-organization, and the growth of international collaboration in science. Research Policy, 34(10), 1608–1618.
Xie, Z., Li, J. P., & Li, M. (2018). Exploring cooperative game mechanisms of scientific coauthorship networks. Complexity,. https://doi.org/10.1155/2018/9173186.
Xie, Z., Li, M., Li, J. P., Duan, X. J., & Ouyang, Z. Z. (2018). Feature analysis of multidisciplinary scientific collaboration patterns based on pnas. EPJ Data Science, 7, 5.
Xie, Z., Ouyang, Z. Z., & Li, J. P. (2016). A geometric graph model for coauthorship networks. Journal of Informetrics, 10, 299–311.
Xie, Z., Ouyang, Z. Z., Li, J. P., Dong, E. M., & Yi, D. Y. (2018). Modelling transition phenomena of scientific coauthorship networks. Journal of the Association for Information Science and Technology, 69(2), 305–317.
Xie, Z., Xie, Z. L., Li, M., Li, J. P., & Yi, D. Y. (2017). Modeling the coevolution between citations and coauthorship of scientific papers. Scientometrics, 112, 483–507.
Zeng, A., Shen, Z., Zhou, J., Wu, J., Fan, Y., Wang, Y., et al. (2017). The science of science: From the perspective of complex systems. Physics Reports, 714, 1–73.
Acknowledgements
The author thinks Professor Jinying Su in the National University of Defense Technology and anonymous reviewers for their helpful comments and feedback. This work is supported by the National Natural Science Foundation of China (Grant No. 61773020).
Author information
Authors and Affiliations
Corresponding author
Appendices
Appendix A
Ambiguities exist in coauthorship data, known as merging and splitting errors. Since there are many nodes with a small degree or hyperdegree, these errors do not change the feature of the heads of degree and hyperdegree distributions, as well as the heads of C(k) and N(k). Merging errors can ruin the exponential cutoffs of degree and hyperdegree distributions, because they would generate nodes with a degree far more than ground truth. Splitting errors generate nodes with a degree smaller than ground truth; thus would generate the exponential cutoffs.
Consider a dataset suffering heavy merging errors and free of splitting errors. If the dataset still has exponential cutoffs in its degree and hyperdegree distributions, we could say these distributions of the corresponding ground-truth dataset have an exponential cutoff. In fact, the density function of the summation of two random variables drawn from an exponential distribution still has an exponential factor. Generate entities through short name (surname and the initial of the first given name) and use them to construct networks (PNAS-s, PRE-s). These networks are almost free of splitting errors (unless an author provides a wrong name), but heavily suffer merging errors. Table 1 shows certain statistical indexes of these networks. Figure 6 shows the degree and hyperdegree distributions still have an exponential cutoff, which means the cutoff can be regarded as a feature of the corresponding ground-truth distributions.
Merging errors satisfying specific distributions can generate a power law. Therefore, our conclusions are drawn based on the assumption that the ground-truth distributions of degrees and hyperdegrees have a power-law part, because the considered data are not free of merging errors. The explanation for the features of the tails of C(k) and N(k) is also dependent on this assumption.
To show the reasonability of this assumption, we computed the proportion of short names in the author names of papers, and that of the short names appeared in more than one paper, because using short names will generate a lot of merging errors. Chinese names were also found to account for merging errors (Kim and Diesner 2016). For the names on papers, we calculated the proportion of the names consisting of a surname among major 100 Chinese surnames and a given name less than six characters. The small proportion of such names and the much small proportion of such names appeared in more than one paper imply the impact of merging errors is limited on the considered datasets, especially on the dataset PNAS (Table 3).
Appendix B
The hyperedge-size distributions can be fitted by a mixture of a generalized Poisson and a power-law distribution. Let the domains of generalized Poisson \(f_1(x)= {a (a+bx)^{x-1}}{ \mathrm {e}^{-a-bx }/{ x!}}\), cross-over, and power-law \(f_2(x)=cx^{-d}\) be \([\min (x),E]\), [B, E], and \([B,\max (x)]\) respectively. The fitting function is
where \(q(x)=\mathrm {e}^{ - (x -B)/(E-x ) }\). The fitting process is: calculate parameters of \(sf_1(x)\) and \(f_2(x)\) by regressing the head and tail of a given distribution respectively; find B and E through exhaustion to make f(x) pass the KS test, namely p-value \(>0.05\). This value larger than 0.05 means the test cannot reject the hypothesis of following the same distribution at significance level \(5\%\). The fitting results are listed in Table 4.
Appendix C
Table 5 shows the boundary detection algorithm for generalized Poisson distributions. The observations \(\{D_s, s=1,\ldots ,n\}\) are nodes’ degree or hyperdegree. The input of the algorithm is \(h(x)=a(a+bx)^{x-1}\mathrm {e}^{-a-bx}/x!\), where \(a, b, x\in {{\mathbb {R}}}\). Changing the algorithm as follows, we can obtain the boundary of power-law distributions with an exponential cutoff. For k from 1 to \(\max (D_1,D_2,\ldots ,D_n)\), we fit the density \(h(x) \propto x^{-\mu }\mathrm {e}^{-{\nu }{x}}\) (where \(\mu , \nu \in {{\mathbb {R}}}\)) to the observations not smaller than k until the KS test accepts the null hypothesis that these observations follow the distribution with density h(x). Table 6 shows the fitting results for the heads and tails of degree and hyperdegree distributions.
Table 7 shows the boundary detection algorithm for the average local clustering coefficient of k-degree nodes C(k) and the average degree of k-degree nodes’ neighbors N(k) (Xie et al. 2018). The inputs are \(g(\cdot )=\log (\cdot )\), \(h(s)=a_1 \mathrm {e}^{-((s-a_2)/a_3)^2}\) for C(k), and \(h(s)= a_1 s^3 + a_2 s^2 + a_3 s + a_4\) for N(k), where s, \(a_i\in {{\mathbb {R}}}\) (\(i=1,\ldots ,4\)). Using these inputs is based on the observations on C(k) and N(k).
Appendix D
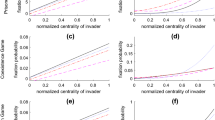
We compared the provided model with two typical models of coauthorship networks (Barabási et al. 2002; Catanzaro et al. 2004), the pseudo codes of which are listed in Tables 8 and 9. The first model captures node clustering, but cannot predict degree assortativity. And the second model captures degree assortativity, but cannot predict node clustering (Table 10). Both of them neglect the aging of nodes, and cannot capture the multimodality of the empirical coauthorship networks (Fig. 7).
Rights and permissions
About this article
Cite this article
Xie, Z. A cooperative game model for the multimodality of coauthorship networks. Scientometrics 121, 503–519 (2019). https://doi.org/10.1007/s11192-019-03183-z
Received:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11192-019-03183-z