Abstract
This paper addresses the problem of information burying in social sciences, where a large amount of experimental findings reported in multiple scientific articles may be missed by scholars due to the lack of an active accumulation, organization and synthesis of these findings into a centralized information system. To tackle the information burying problem, in this paper we present a new network-based data model and methodology for aggregating, organizing, linking and mining quantitative results published in multiple academic articles in particular sub-fields of social sciences. The goal of the proposed methodology is to provide researchers with a wider perspective when viewing scientific results in their own fields and utilize it for their research. To validate the proposed approach, we conducted a manual experiment with a corpus of 41 scientific articles in the field of personal information management. The experiment indicates that the constructed network-based information system can be effectively used to explore the relationships between the results of various articles, raising new research questions and hypotheses based on results from multiple articles that tested similar variables. The proposed system can serve as a catalyst for the advancement of research in various fields of social science.



Similar content being viewed by others
Notes
National Science Board (2018). Science and engineering indicators 2018. Arlington, VA: National Science Foundation. Retrieved from https://www.nsf.gov/statistics/2018/nsb20181/report/sections/academic-research-and-development/outputs-of-s-e-research-publications.
National Science Board (2016). Science and engineering indicators 2016. Arlington, VA: National Science Foundation. Retrieved from https://www.nsf.gov/statistics/2016/nsb20161/uploads/1/nsb20161.pdf.
Retrieved from http://www.w3.org/TR/rdf-sparql-query/.
References
Bechhofer, S., Buchan, I., De Roure, D., Missier, P., Ainsworth, J., Bhagat, J., et al. (2013). Why linked data is not enough for scientists. Future Generation Computer Systems,29(2), 599–611.
Bergman, O., & Whittaker, S. (2016). The science of managing our digital stuff. Cambridge, MA: MIT Press.
Blondel, V. D., Guillaume, J.-L., Lambiotte, R., & Lefebvre, E. (2008). Fast unfolding of communities in large networks. Journal of Statistical Mechanics: Theory and Experiment, 2008(10), P10008.
Bonacich, P. (1987). Power and centrality: A family of measures. American Journal of Sociology,92(5), 1170–1182.
Borenstein, M., Hedges, L. V., Higgins, Julian P. T., & Rothstein, H. R. (2009). Introduction to meta-analysis. Hoboken, NJ: Wiley.
Borgatti, S. P. (2005). Centrality and network flow. Social Networks,27(1), 55–71.
Brown, P. O., & Botstein, D. (1999). Exploring the new world of the genome with DNA microarrays. Nature Genetics,21(1s), 33.
Casillas, L., & Daradoumis, T. (2012). An ontological structure for gathering and sharing knowledge among scientists through experiment modeling. In Collaborative and distributed E-research: Innovations in technologies, strategies and applications (pp. 165–179). IGI Global.
Cheadle, C., Cao, H., Kalinin, A., & Hodgkinson, J. (2017). Advanced literature analysis in a Big Data world. Annals of the New York Academy of Sciences,1387(1), 25–33.
Chen, L., & Friedman, C. (2004). Extracting phenotypic information from the literature via natural language processing (pp. 758–762). San-Francisco: Medinfo.
Ciccarese, P., Elizabeth, W., Wong, G., Ocana, M., Kinoshita, J., Ruttenberg, A., et al. (2008). The SWAN biomedical discourse ontology. Journal of Biomedical Informatics,41(5), 739–751.
De Roure, D., Goble, C., Aleksejevs, S., Bechhofer, S., Bhagat, J., Cruickshank, D., et al. (2010). The evolution of myexperiment. In 2010 IEEE Sixth International Conference on e-Science (e-Science).
Etzioni, O., Banko, M., Soderland, S., & Weld, D. S. (2008). Open information extraction from the web. Communications of the ACM,51(12), 68–74.
Feichtinger, J., McFarlane, R. J., & Larcombe, L. D. (2012). CancerMA: A web-based tool for automatic meta-analysis of public cancer microarray data. Database 2012.
Fiszman, M., Demner-Fushman, D., Kilicoglu, H., & Rindflesch, T. C. (2009). Automatic summarization of MEDLINE citations for evidence-based medical treatment: A topic-oriented evaluation. Journal of Biomedical Informatics,42(5), 801–813.
Friedman, C., Kra, P., Yu, H., Krauthammer, M., & Rzhetsky, A. (2001). GENIES: A natural-language processing system for the extraction of molecular pathways from journal articles. In Proceedings of ISMB (supplement of bioinformatics) conference, Copenhagen, Denmark (pp. 74–82).
Gruber, T. R. (1995). Toward principles for the design of ontologies used for knowledge sharing? International Journal of Human-Computer Studies,43(5–6), 907–928.
Hearst, M. A. (1992). Automatic acquisition of hyponyms from large text corpora. In Proceedings of the 14th conference on computational linguistics (Vol. 2).
Hearst, M. A. (2006). Clustering versus faceted categories for information exploration. Communications of the ACM,49(4), 59–61. https://doi.org/10.1145/1121949.1121983.
Henderson, J., & Popa, D. N. (2016). A vector space for distributional semantics for entailment. arXiv preprint arXiv:1607.03780.
Higgins, J. P. T., & Green, S. (2005). Cochrane handbook for systematic reviews of interventions. Version.
Holzinger, A., Simonic, K.-M., & Yildirim, P. (2012). Disease–disease relationships for rheumatic diseases: Web-based biomedical textmining an knowledge discovery to assist medical decision making. In 2012 IEEE 36th annual computer software and applications conference.
Jankowski, N. W. (2007). Exploring e-science: An introduction. Journal of Computer-Mediated Communication,12(2), 549–562.
Keshtkaran, A., Yuhaniz, S. S., & Ibrahim, S. (2017). An overview of cross-document coreference resolution. In 2017 international conference on computer and drone applications (IConDA).
Kotlerman, L., Dagan, I., Szpektor, I., & Zhitomirsky-Geffet, M. (2010). Directional distributional similarity for lexical inference. Natural Language Engineering,16(4), 359–389.
Kozareva, Z., & Hovy, E. (2010). A semi-supervised method to learn and construct taxonomies using the web. In Proceedings of the 2010 conference on empirical methods in natural language processing.
Krallinger, M., Leitner, F., Rabal, O., Vazquez, M., Oyarzabal, J., & Valencia, A. (2015). CHEMDNER: The drugs and chemical names extraction challenge. Journal of Cheminformatics,7(1), S1.
Krauthammer, M., & Nenadic, G. (2004). Term identification in the biomedical literature. Journal of Biomedical Informatics,37(6), 512–526.
Lapesa, G., Kawaletz, L., Plag, I., Andreou, M., Kisselew, M., & Pado, S. (2018). Disambiguation of newly derived nominalizations in context: A distributional semantics approach. Word Structure, 11(3), 277–312.
Larsen, K. R., & Bong, C. H. (2016). A tool for addressing construct identity in literature reviews and meta-analyses. MIS Quarterly,40(3), 529–551.
Liu, Y., Bill, R., Fiszman, M., Rindflesch, T., Pedersen, T., Melton, G. B., et al. (2012). Using SemRep to label semantic relations extracted from clinical text. In AMIA annual symposium proceedings.
Liu, K., Hogan, W. R., & Crowley, R. S. (2011). Natural language processing methods and systems for biomedical ontology learning. Journal of Biomedical Informatics,44(1), 163–179.
Manning, C. D., & Schütze, H. (1999). Foundations of statistical natural language processing (Vol. 999). Cambridge: MIT Press.
McGuinness, D. L., & Van Harmelen, F. (2004). OWL web ontology language overview. W3C Recommendation,10(10), 2004.
Mikolov, T., Chen, K., Corrado, G., & Dean, J. (2013). Efficient estimation of word representations in vector space. arXiv preprint arXiv:1301.3781.
Mirkin, S., Dagan, I., & Geffet, M. (2006). Integrating pattern-based and distributional similarity methods for lexical entailment acquisition. In Proceedings of the COLING/ACL on Main conference poster sessions.
Mohri, M., Rostamizadeh, A., & Talwalkar, A. (2012). Foundations of machine learning. Cambridge: MIT Press.
Mons, B. (2005). Which gene did you mean? BMC Bioinformatics,6(1), 142.
Moretti, F. (2005). Graphs, maps, trees: Abstract models for a literary history. London: Verso.
Mueller, R., & Abdullaev, S. (2019). DeepCause: Hypothesis extraction from information systems papers with deep learning for theory ontology learning. In Proceedings of the 52nd Hawaii international conference on system sciences.
Mueller, R. M., & Huettemann, S. (2018). Extracting causal claims from information systems papers with natural language processing for theory ontology learning. In Proceedings of the 51st Hawaii international conference on system sciences.
Nickel, M., Murphy, K., Tresp, V., & Gabrilovich, E. (2016). A review of relational machine learning for knowledge graphs. Proceedings of the IEEE,104(1), 11–33.
Noy, N. F, & McGuinness, D. L. (2001). Ontology development 101: A guide to creating your first ontology. Stanford knowledge systems laboratory technical report KSL-01-05 and Stanford medical informatics technical report SMI-2001-0880, Stanford, CA.
Panchenko, A., Faralli, S., Ruppert, E., Remus, S., Naets, H., Fairon, C., et al. (2016). TAXI at SemEval-2016 Task 13: A taxonomy induction method based on lexico-syntactic patterns, substrings and focused crawling. In Proceedings of the 10th international workshop on semantic evaluation (SemEval-2016).
Polhill, J. G., Pignotti, E., Gotts, N. M., Edwards, P., & Preece, A. (2007). A semantic grid service for experimentation with an agent-based model of land-use change. Journal of Artificial Societies and Social Simulation,10(2), 2.
Pontika, N., Knoth, P., Cancellieri, M., & Pearce, S. (2015). Fostering open science to research using a taxonomy and an eLearning portal. In Proceedings of the 15th international conference on knowledge technologies and data-driven business.
Rinaldi, F., Schneider, G., Kaljurand, K., Hess, M., Andronis, C., Konstandi, O., et al. (2007). Mining of relations between proteins over biomedical scientific literature using a deep-linguistic approach. Artificial Intelligence in Medicine,39(2), 127–136.
Segura-Bedmar, I., Suárez-Paniagua, V., & Martínez, P. (2015). Exploring word embedding for drug name recognition. In Proceedings of the sixth international workshop on health text mining and information analysis.
Stern, A., & Dagan, I. (2014). Recognizing implied predicate–argument relationships in textual inference. In Proceedings of the 52nd annual meeting of the association for computational linguistics (Vol. 2: Short Papers).
Tchoua, R. B., Chard, K., Audus, D. J., Ward, L. T., Lequieu, J., De Pablo, J. J., & Foster, I. T. (2017). Towards a hybrid human-computer scientific information extraction pipeline. In 2017 IEEE 13th international conference on e-Science (e-Science).
Tsoi, L. C., Patel, R., Zhao, W., & Zheng, W. J. (2009). Text-mining approach to evaluate terms for ontology development. Journal of Biomedical Informatics,42(5), 824–830.
Vandenbroeck, P., Goossens, J., & Clemens, M. (2007). Tackling obesities: Future choices—obesity system Atlas. edited by Government Office for Science. London.
Wities, R., Shwartz, V., Stanovsky, G., Adler, M., Shapira, O., Upadhyay, S., et al. (2017). A consolidated open knowledge representation for multiple texts. In Proceedings of the 2nd workshop on linking models of lexical, sentential and discourse-level semantics.
Xu, C., Cao, H., Zhang, F., & Cheadle, C. (2018). Comprehensive literature data-mining analysis reveals a broad genetic network functionally associated with autism spectrum disorder. International Journal of Molecular Medicine, 42(5), 2353–2362.
Zhou, D., & He, Y. (2008). Extracting interactions between proteins from the literature. Journal of Biomedical Informatics,41(2), 393–407.
Acknowledgements
We thank our research assistants Natalie Friedman and Sarah Cohen for their excellent work. This study was partly supported by Google Faculty Research Award 2014_R2_79.1.
Author information
Authors and Affiliations
Corresponding author
Additional information
This paper is dedicated to the memory of Judit Bar-Ilan (1958–2019), an outstanding scholar and an inimitable friend and colleague.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Appendix
Appendix
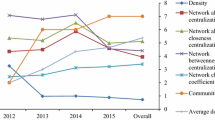
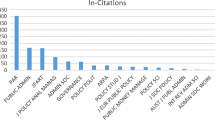
See Tables 1, 2, 3 and 4 and Figs. 4, 5, 6, 7, 8 and 9.
Rights and permissions
About this article
Cite this article
Zhitomirsky-Geffet, M., Bergman, O. & Hilel, S. Towards a wider perspective in the social sciences using a network of variables based on thousands of results. Scientometrics 123, 1385–1406 (2020). https://doi.org/10.1007/s11192-020-03446-0
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11192-020-03446-0