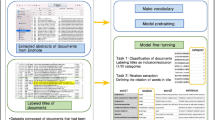
Abstract
Document screening is a fundamental task within Evidence-based Medicine (EBM), a practice that provides scientific evidence to support medical decisions. Several approaches have tried to reduce physicians’ workload of screening and labeling vast amounts of documents to answer clinical questions. Previous works tried to semi-automate document screening, reporting promising results, but their evaluation was conducted on small datasets, which hinders generalization. Moreover, recent works in natural language processing have introduced neural language models, but none have compared their performance in EBM. In this paper, we evaluate the impact of several document representations such as TF-IDF along with neural language models (BioBERT, BERT, Word2Vec, and GloVe) on an active learning-based setting for document screening in EBM. Our goal is to reduce the number of documents that physicians need to label to answer clinical questions. We evaluate these methods using both a small challenging dataset (CLEF eHealth 2017) as well as a larger one but easier to rank (Epistemonikos). Our results indicate that word as well as textual neural embeddings always outperform the traditional TF-IDF representation. When comparing among neural and textual embeddings, in the CLEF eHealth dataset the models BERT and BioBERT yielded the best results. On the larger dataset, Epistemonikos, Word2Vec and BERT were the most competitive, showing that BERT was the most consistent model across different corpuses. In terms of active learning, an uncertainty sampling strategy combined with a logistic regression achieved the best performance overall, above other methods under evaluation, and in fewer iterations. Finally, we compared the results of evaluating our best models, trained using active learning, with other authors methods from CLEF eHealth, showing better results in terms of work saved for physicians in the document-screening task.














Similar content being viewed by others
Notes
References
Adeva, J. G., Atxa, J. P., Carrillo, M. U., & Zengotitabengoa, E. A. (2014). Automatic text classification to support systematic reviews in medicine. Expert Systems with Applications, 41(4), 1498–1508.
Alharbi, A., Briggs, W., & Stevenson, M. (2018). Retrieving and ranking studies for systematic reviews: University of sheffield’s approach to CLEF eHealth 2018 task 2. In CEUR workshop proceedings (Vol. 2125).
Alharbi, A., & Stevenson, M. (2017). Ranking abstracts to identify relevant evidence for systematic reviews: The university of sheffield’s approach to CLEF eHealth 2017 task 2. In Clef (working notes).
Alharbi, A., & Stevenson, M. (2019). Ranking studies for systematic reviews using query adaptation: University of sheffield’s approach to CLEF eHealth 2019 task 2. In Clef (working notes).
Bekhuis, T., Tseytlin, E., Mitchell, K. J., & Demner-Fushman, D. (2014). Feature engineering and a proposed decision-support system for systematic reviewers of medical evidence. PloS ONE, 9(1), e86277.
Chen, J., Chen, S., Song, Y., Liu, H., Wang, Y., Hu, Q., & Yang, Y. (2017). ECNU at 2017 eHealth task 2: Technologically assisted reviews in empirical medicine. In CLEF (working notes).
Choi, S., Ryu, B., Yoo, S., & Choi, J. (2012). Combining relevancy and methodological quality into a single ranking for evidence-based medicine. Information Sciences, 21(4), 76–90.
Cohen, A. M., & Smalheiser, N. R. (2018). UIC/OHSU CLEF 2018 task 2 diagnostic test accuracy ranking using publication type cluster similarity measures. In CEUR workshop proceedings (Vol. 2125).
Cormack, G. V., & Grossman, M. R. (2016). “ when to stop” waterloo (CORMACK) participation in the TREC 2016 total recall track. In TREC.
Cormack, G. V., & Grossman, M. R. (2017). Technology-assisted review in empirical medicine: Waterloo participation in CLEF eHealth 2017. In CLEF (working notes).
Dehghani, M., Zamani, H., Severyn, A., Kamps, J., & Croft, W. B. (2017). Neural ranking models with weak supervision. In Proceedings of the 40th international ACM SIGIR conference on research and development in information retrieval (pp. 65–74).
Del Fiol, G., Michelson, M., Iorio, A., Cotoi, C., & Haynes, R. B. (2018). A deep learning method to automatically identify reports of scientifically rigorous clinical research from the biomedical literature: Comparative analytic study. Journal of Medical Internet Research, 20(6), 10281.
Devlin, J., Chang, M.W., Lee, K., & Toutanova, K. (2018). Bert: pre-training of deep bidirectional transformers for language understanding. arXiv:1810.04805.
Di Nunzio, G. M. (2019). A distributed effort approach for systematic reviews. IMS UNIPD at CLEF 2019 eHealth task 2. In Clef (working notes).
Di Nunzio, G. M., Beghini, F., Vezzani, F., & Henrot, G. (2017). An interactive two-dimensional approach to query aspects rewriting in systematic reviews. IMS UNIPD at CLEF eHealth task 2. In CLEF (working notes).
Di Nunzio, G. M., Ciuffreda, G., & Vezzani, F. (2018). Interactive sampling for systematic reviews. IMS UNIPD at CLEF 2018 eHealth task 2. In CLEF (working notes).
Donoso-Guzmán, I., & Parra, D. (2018). An interactive relevance feedback interface for evidence-based health care. In 23rd international conference on intelligent user interfaces (pp. 103–114).
Elliott, J. H., Turner, T., Clavisi, O., Thomas, J., Higgins, J. P., Mavergames, C., et al. (2014). Living systematic reviews: An emerging opportunity to narrow the evidence-practice gap. PLoS Medicine, 11(2), e1001603.
Figueroa, R. L., Zeng-Treitler, Q., Ngo, L. H., Goryachev, S., & Wiechmann, E. P. (2012). Active learning for clinical text classification: is it better than random sampling? Journal of the American Medical Informatics Association, 19(5), 809–816.
Goeuriot, L., Kelly, L., Suominen, H., Névéol, A., Robert, A., Kanoulas, E., & Zuccon, G. (2017). CLEF 2017 eHealth evaluation lab overview. In International conference of the cross-language evaluation forum for European languages (pp. 291–303).
Goodwin, T. R., & Harabagiu, S. M. (2018). Knowledge representations and inference techniques for medical question answering. In ACM transactions on intelligent systems and technology (TIST) 9214 .
Grossman, M. R., Cormack, G. V., & Roegiest, A. (2016). Trec 2016 total recall track overview. In TREC.
Hashimoto, K., Kontonatsios, G., Miwa, M., & Ananiadou, S. (2016). Topic detection using paragraph vectors to support active learning in systematic reviews. Journal of Biomedical Informatics, 6(2), 59–65.
Hollmann, N., & Eickhoff, C. (2017). Ranking and feedback-based stopping for recall-centric document retrieval. In CLEF (working notes).
Howard, J., & Ruder, S. (2018). Universal language model fine-tuning for text classification. arXiv:1801.06146.
Hughes, M., Li, I., Kotoulas, S., & Suzumura, T. (2017). Medical text classification using convolutional neural networks. Stud Health Technol Inform, 235, 246–50.
Joachims, T. (1998). Text categorization with support vector machines: Learning with many relevant features. In European conference on machine learning (pp. 137–142).
Kalphov, V., Georgiadis, G., & Azzopardi, L. (2017). Sis at CLEF 2017 eHealth tar task. In CEUR workshop proceedings (Vol. 1866, pp. 1–5).
Kanoulas, E., Li, D., Azzopardi, L., & Spijker, R. (2017). CLEF 2017 technologically assisted reviews in empirical medicine overview. In CEUR workshop proceedings (Vol. 1866, pp. 1–29).
Kanoulas, E., Li, D., Azzopardi, L., & Spijker, R. (2018). CLEF 2018 technologically assisted reviews in empirical medicine overview. CEUR workshop proceedings (Vol. 1866, pp. 1–34).
Kanoulas, E., Li, D., Azzopardi, L., & Spijker, R. (2019). CLEF 2019 technology assisted reviews in empirical medicine overview. In CEUR workshop proceedings (Vol. 2380).
Keselman, A., & Smith, C. A. (2012). A classification of errors in lay comprehension of medical documents. Journal of Biomedical Informatics, 45(6), 1151–1163.
Lagopoulos, A., Anagnostou, A., Minas, A., & Tsoumakas, G. (2018). Learning-to-rank and relevance feedback for literature appraisal in empirical medicine. In International conference of the cross-language evaluation forum for European languages (pp. 52–63).
Lee, G. E. (2017). A study of convolutional neural networks for clinical document classification in systematic reviews: Sysreview at CLEF eHealth 2017.
Lee, G. E., & Sun, A. (2018). Seed-driven document ranking for systematic reviews in evidence-based medicine. In The 41st international ACM SIGIR conference on research & development in information retrieval (pp. 455–464).
Lee, J., Yoon, W., Kim, S., Kim, D., Kim, S., So, C. H., & Kang, J. (2019). Biobert: pre-trained biomedical language representation model for biomedical text mining. arXiv:1901.08746.
Li, D., Kanoulas, E. et al. (2019). Automatic thresholding by sampling documents and estimating recall: Ilps@ uva at tar task 2.2. In CEUR workshop proceedings (Vol. 2380).
Mikolov, T., Sutskever, I., Chen, K., Corrado, G. S., & Dean, J. (2013). Distributed representations of words and phrases and their compositionality. In Advances in neural information processing systems (pp. 3111–3119).
Minas, A., Lagopoulos, A., & Tsoumakas, G. (2018). Aristotle university’s approach to the technologically assisted reviews in empirical medicine task of the 2018 CLEF eHealth lab. In CLEF (working notes).
Miwa, M., Thomas, J., O’Mara-Eves, A., & Ananiadou, S. (2014). Reducing systematic review workload through certainty-based screening. Journal of Biomedical Informatics, 51, 242–253.
Mo, Y., Kontonatsios, G., & Ananiadou, S. (2015). Supporting systematic reviews using LDA-based document representations. Systematic Reviews, 4(1), 172.
Nogueira, R., Yang, W., Cho, K., & Lin,J. (2019). Multi-stage document ranking with BERT. arXiv:1910.14424.
Norman, C., Leeflang, M., & Névéol, A. (2017). Limsi@ CLEF eHealth 2017 task 2: Logistic regression for automatic article ranking.
Norman, C. R., Leeflang, M. M., & Névéol, A. (2018). Limsi@ CLEF eHealth 2018 task 2: Technology assisted reviews by stacking active and static learning. In CLEF (working notes) 2125 (pp. 1–13).
Pedregosa, F., Varoquaux, G., Gramfort, A., Michel, V., Thirion, B., & Grisel, O. (2011). Scikit-learn: Machine learning in python. Journal of Machine Learning Research, 12, 2825–2830.
Pennington, J., Socher, R., & Manning, C. (2014). Glove: Global vectors for word representation. In: Proceedings of the 2014 conference on empirical methods in natural language processing (EMNLP) (pp. 1532–1543).
Peters, M. E., Neumann, M., Iyyer, M., Gardner, M., Clark, C., Lee, K., & Zettlemoyer, L. (2018). Deep contextualized word representations. arXiv:1802.05365.
Qiao, Y., Xiong, C., Liu, Z., & Liu, Z. (2019). Understanding the behaviors of bert in ranking. arXiv:1904.07531.
Ratner, A., Bach, S. H., Ehrenberg, H., Fries, J., Wu, S., & Ré, C. (2017). Snorkel: Rapid training data creation with weak supervision. Proceedings of the VLDB Endowment, 11(3), 269–282.
Roy, D., Ganguly, D., Bhatia, S., Bedathur, S., & Mitra, M. (2018). Using word embeddings for information retrieval: How collection and term normalization choices affect performance. In Proceedings of the 27th ACM international conference on information and knowledge management (pp. 1835–1838).
Salton, G., Wong, A., & Yang, C. S. (1975). A vector space model for automatic indexing. Communications of the ACM, 18(11), 613–620.
Scells, H., Zuccon, G., Deacon, A., & Koopman, B. (2017). Qut IELAB at CLEF eHealth 2017 technology assisted reviews track: initial experiments with learning to rank. In CEUR workshop proceedings: Working notes of CLEF 2017: Conference and labs of the evaluation forum (Vol. 1866, pp. Paper–98).
Settles, B. (2012). Active learning. Synthesis Lectures on Artificial Intelligence and Machine, Learning, 6(1), 1–114.
Singh, G., Marshall, I., Thomas, J., & Wallace, B. (2017). Identifying diagnostic test accuracy publications using a deep model. In CEUR workshop proceedings (Vol. 1866).
Singh, J., & Thomas, L. (2017). Iiit-h at CLEF eHealth 2017 task 2: Technologically assisted reviews in empirical medicine. In CLEF (working notes).
van Altena, A. J., & Olabarriaga, S. D. (2017). Predicting publication inclusion for diagnostic accuracy test reviews using random forests and topic modelling. In CLEF (working notes).
Vaswani, A., Shazeer, N., Parmar, N., Uszkoreit, J., Jones, L., Gomez, A. N., & Polosukhin, I. (2017). Attention is all you need. In Advances in neural information processing systems (pp. 5998–6008).
Wallace, B. C., Small, K., Brodley, C. E., Lau, J., Schmid, C. H., Bertram, L., et al. (2012). Toward modernizing the systematic review pipeline in genetics: Efficient updating via data mining. Genetics in Medicine, 14(7), 663.
Wallace, B. C., Small, K., Brodley, C. E., & Trikalinos, T. A. (2010). Active learning for biomedical citation screening. In Proceedings of the 16th ACM SIGKDD international conference on knowledge discovery and data mining (pp. 173–182).
Wu, H., Wang, T., Chen, J., Chen, S., Hu, Q., & He, L. (2018). ECNU at 2018 eHealth task 2: Technologically assisted reviews in empirical medicine. Methods, 4(5), 7.
Yang, Y. Y., Lee, S. C., Chung, Y. A., Wu, T. E., Chen, S. A., & Lin, H. T. (2017). LIBACT: Pool-based active learning in python.
Yu, Z., & Menzies, T. (2017). Data balancing for technologically assisted reviews: Undersampling or reweighting. In CLEF (working notes).
Acknowledgments
This research was funded by ANID Chile, Fondecyt Grant 1191791, and the Millenium Institute Foundational Research on Data (IMFD).
Author information
Authors and Affiliations
Corresponding author
Appendix
Appendix
Appendix 1: Leave one out cross-validation CLEF eHealth 2017
In this section we do a leave one out cross-validation evaluation. Concerning model input we use the same method to represent documents and medical questions pairs representation proposed in “Comparison with participants from CLEF eHealth challenge” section, allowing it to be possible to use this evaluation methodology. Now, instead of having one model for each medical question, we train a general model to make predictions for new questions and then rank the documents. We evaluate the performance of our active learning framework using leave one out cross-validation for the 50 queries included in the CLEF eHealth 2017 task 2 dataset on our four best combinations of active learning strategies, machine learning model, and language model representations (US-BioBERT-RF, US-BERT-RF, US-BioBERT-LR, and US-BERT-LR). The evaluation metrics used were the average of each medical question testing and training with the rest on the last relevant document found (lastrel), work saved oversampling (wss95 and wss100), average precision (ap), and normalized cumulative gain at recall@k% (20,40,60).
For carrying out this evaluation, we compare the leave one out cross-validation on our best four models obtained from experiments made in “Results” section for the CLEF eHealth 2017 task 2 dataset. We train with all the questions and test with one, repeating this process iteratively until we have evaluation results for each medical query. Regarding the metrics used for the evaluation, we used parameters related to saved work (wss100 and wss95), accumulated recall (ncg20,ncg40, and ncg60), and precision (ap).
The results on Table 7 show that in terms of the last relevant document (lastrel), the method US-RF-BIOBERT is the best among our other models. With these results, we confirm that this is our best model for the task of retrieving all the relevant evidence given a medical question. More evidence of this effect is provided by work-saved oversampling (wss100 and wss95), which indicates that our best approach allows the physician to save near 60% of their work. Then, concerning cumulative gain (ncg20, ncg40, ncg60), US-BioBERT-RF ranks a 92% of the relevant documents on the first 60% of the candidates list. Finally, in terms of precision, US-RF-BIOBERT obtains the best results among the other models; however, it finds only 18.2 percent of the relevant documents over the total of retrieved ones.
Appendix 2: CLEF eHealth 2017 task 2 test dataset
Appendix 3: Epistemonikos dataset
Rights and permissions
About this article
Cite this article
Carvallo, A., Parra, D., Lobel, H. et al. Automatic document screening of medical literature using word and text embeddings in an active learning setting. Scientometrics 125, 3047–3084 (2020). https://doi.org/10.1007/s11192-020-03648-6
Received:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11192-020-03648-6