Abstract
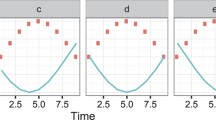
Microarray technologies are enabling measurements of gene expression levels at large scales. Applications of microarrays in medicine, biological engineering and agriculture often depend on quantitative studies of time-series data. For drug delivery, fed batch reactors and crop growth, time series analysis is essential in optimizing controlled biological processes. In particular, it is useful to project discrete time series data into continuous dynamic trajectories with a global gene expression model. Time series microarray data were fitted with differential equations based on Singular Value Decomposition. Using the data from two different organisms, Saccharomyces Cerevisiae and Drosophila melanogaster the local predictive accuracy was assessed with cross-fold validation across all experiments. The equations were integrated and inspected visually. The algorithm for inducing differential equations was found to produce a good fit both globally and locally.
Similar content being viewed by others
References
G. Stephanopoulos, A. Aristidou, and J. Nielsen, “Metabolic Engineering, Principles and Methodologies,” Academic, 1998. pp. 309–351, 411–459.
E.M Blalock (Ed.), “A Beginner’s Guide to Microarrays,” Kluwer, 2003.
P. Baldi and W.G. Hatfield, “DNA Microarrays and Gene Expression,” Cambridge University Press, 2002.
H. Liu, S. Tarima, A.S. Borders, T.V. Getchell, M.L. Getchell, and A.J. Stromberg, “Quadratic Regression Analysis for Gene Discovery and Pattern Recognition for Non-cyclic Short Time-course Microarray Experiments,” BMC Bioinformatics, vol. 6, 2005, p. 106.
P. Ma, C.I. Castillo-Davis, W. Zhong, and J.S. Liu, “A Data-Driven Clustering Method for Time Course Gene Expression Data,” Nucleic Acids Res. vol. 34, no. 4, 2006, pp. 1261–1269, (Mar 1).
R. Opgen-Rhein and K. Strimmer, “Learning Causal Networks from Systems Biology Time Course Data: An Effective Model Selection Procedure for Vector Autoregressive Process,” BMC Bioinformatics, vol. 8, 2007, (in press).
S. Wichert, K. Fokianos and K. Strimmer, “Identifying Periodically Expressed Transcripts in Microarray Time Series Data,” Bioinformatics, vol. 20, 2004, pp. 5–20.
O. Alter, P. Brown and D. Botstein, “Singular Value Decomposition for Genome-wide Expression Data Processing And Modeling,” Genetics, vol. 97, no. 18, 2000, pp. 10101–10106.
M. Yeung, J. Tegner and J. Collins, “Reverse Engineering Gene Networks Using Singular Value Decomposition and Robust Regression,” Genetics, vol. 99, no. 9, 2002, pp. 6161–6168.
X. Leng and H. Muller, “Classification Using Functional Data Analysis for Temporal Gene Expression Data, Bioinformatics, vol. 22, no. 1, 2006, pp. 68–76.
K. Wright, “Differential Equations for the Analytic Singular Value Decomposition of a Matrix,” Numerische Math., vol. 63, pp. 283–295.
J. Gollub, C. Ball, G. Binkley, J. Demeter, D. Finkelstein, J. Hebert, T. Hernandez-Boussard, H. Jin, M. Kaloper, J. Matese, M. Schroeder, P. Brown, D. Botstein, and G. Sherlock, “The Stanford Microarray Database: Data Access and Quality Assessment Tools,” Nucleic Acids Res., vol. 31, no. 1, 2003, pp. 94–96.
P. Spellman, G. Sherlock, M. Zhang, V. Iyer, K. Anders, M. Eisen, P. Brown, B. Botstein, and B. Futcher, “Comprehensive Identification of Cell Cycle-regulated Genes of the Yeast Saccharomyces Cerevisiae by Microarray Hybridization,” Mol. Biol. Cell, vol. 9, 1998, pp. 3273–3297.
M. Arbeitman, E. Furlong, F. Imam, E. Johnson, B. Null, B. Baker, M. Krasnow, M. Scott, R. Davis and K. White, “Gene Expression During the Life Cycle of Drosphila melanogaster,” Science, vol. 297, 2002, p. 2270.
W. Press, S. Teukolsky, W. Vetterling and B. Flannery, “Numerical Recipes in C, Second Edition,” Cambridge University Press, 1992, pp. 59–71, 671–680.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Kramer, R., Xu, D. Projecting Gene Expression Trajectories through Inducing Differential Equations from Microarray Time Series Experiments. J Sign Process Syst Sign Image 50, 321–329 (2008). https://doi.org/10.1007/s11265-007-0122-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11265-007-0122-1