Abstract
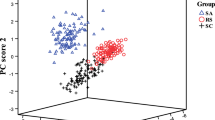
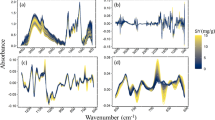
“Sweating” is a traditional processing method of Chinese medicinal materials in place of production. Because this operation is time-consuming, “sweating” is often abandoned, which affects the quality of Chinese medicine. At present, there is no specific method for identification of crude and sweated herbs. In this research, we tried to explore a rapid and effective discrimination method, in order to provide a new means for the quality control of Chinese medicine. We collected 120 batches of data of crude and sweated Chinese medicinal material Dipsaci Radix by near infrared spectroscopy. Support vector machine (SVM) and random forest (RF) were applied to construct discriminant models respectively. For comparison, principal component analysis (PCA)-mahalanobis distance (MD) discriminant was performed. Three models could well classify the test samples with the same error rate 3.33%. However the mean error rates were 1.73% for SVM, 1.13% for RF, and 9.18% for PCA-MD. Meanwhile, RF spent 1.06 s, the shortest time, in computation. Therefore, the performance of RF model is the best for discrimination of crude and sweated Dipsaci Radix.




Similar content being viewed by others
References
Duan, J. A., Su, S. L., Lv, J. L., et al. (2009). Traditional experiences and modern cognition on primary processing of traditional Chinese medicinal materials. Chinese Journal of Chinese Materia Medica, 34(24), 3151–3157.
Duan, J. A., Su, S. L., Yan, H., et al. (2013). “Sweating” of traditional Chinese medicinal materials during primary processing and its mechanisms of enzymatic reaction and chemical conversion. Chinese Traditional and Herbal Drugs, 44(10), 1219–1225.
Zhao, Z., Liang, Z., Chan, K., et al. (2010). A unique issue in the standardization of Chinese Materia Medica: processing. Planta Medica, 76(17), 1975–1986.
Du, W. F., Jiang, D. J., Wu, Y., et al. (2016). Simultaneous determination of isochlorogenic acid A, sochlorogenic acid B, and sochlorogenic acid C in crude and sweated Dipsaci Radix by HPLC. Chinese Journal of Pharmaceutical Analysis, 36(5), 842–846.
Du, W. F., Jia, Y. Q., Jiang, D. J., et al. (2014). Component analysis of crude and sweated Dipsaci Radix based on HPLC-ESI-MS. Chinese Traditional and Herbal Drugs, 45(22), 3251–3255.
Liu, H. L., Yan, R. Y., Guo, J., et al. (2013). Numerical study on the difference of color and odor of Cortex Magnoliae Officinalis before and after sweated. China Journal of Chinese Materia Medica, 38(1), 45–48.
Xu, G. T., Yuan, H. F., & Lu, W. Z. (2000). Development of modern near infrared spectroscopic techniques and its application. Spectroscopy and Spectral Analysis, 20(2), 134–142.
Ding, J. X., Zhang, Q. H., Li, S. L., et al. (2016). Fast detection of glucose and fructose content in honey using NIR spectroscopy. Spectroscopy and Spectral Analysis, 36(10), 197–198.
Ni, L. J., Luan, S. R., & Zhang, L. G. (2016). Exploration of rapidly determining quality of traditional Chinese medicines by (NIR) spectroscopy based on Internet sharing mode. China Journal of Chinese Materia Medica, 41(19), 3520–3527.
Zhu, X. L., Yuan, H. F., & Lu, W. Z. (2004). Progress and application of spectral data pretreatment and wavelength selection methods in NIR analytical technique. Progress in Chemistry, 16(4), 528–542.
Wang, H. Y., Li, J. H., & Yang, F. L. (2014). Overview of support vector machine analysis and algorithm. Application Research of Computers, 31(5), 1281–1286.
Manevitz, L. M., & Yousef, M. (2001). One-class SVMs for document classification. Journal of Machine Learning Research, 2(1), 139–154.
Liu, W. J., Li, W. J., Li, H. G., et al. (2017). Research on the method of identifying maize haploid based on KPCA and near infrared spectrum. Spectroscopy and Spectral Analysis, 37(7), 2024–2027.
Rong, H. N., Zhang, G. X., & Jin, W. D. (2006). Selection of kernel function and parameters for support vector machines in system identification. Journal of System Simulation, 18(11), 3204–3208.
Fanelli, G., Dantone, M., Gall, J., et al. (2013). Random forests for real time 3D face analysis. International Journal of Computer Vision, 101(3), 437–458.
Singh, K., Guntuku, S. C., Thakur, A., et al. (2017). Big data analytics framework for peer-to-peer botnet detection using random forests. Information Sciences, 278(19), 488–497.
Zheng, T. T., Yang, M., Que, Z. J., et al. (2017). Optimization of formula for the proliferation of lung cancer cell line based on random forest regression model. Chinese Journal of Experimental Traditional Medical Formulae, 23(4), 177–182.
Du, W. F., Jia, Y. Q., Jiang, D. J., et al. (2014). Rapid identification analysis of crude and sweated Dipsaci Radix based on near infrared spectroscopy combined with the principal component analysis Mahalanobis distance method. China Journal of Chinese Materia Medica, 39(23), 4603–4607.
Xin, N., Gu, X. F., Wu, H., et al. (2012). Discrimination of raw and processed Dipsacus asperoides, by near infrared spectroscopy combined with least squares-support vector machine and random forests. Spectrochim Acta A Mol Biomol Spectrosc, 89(4), 18–24.
Rodriguez-Galiano, V., Sanchez-Castillo, M., Chica-Olmo, M., et al. (2015). Machine learning predictive models for mineral prospectivity: An evaluation of neural networks, random forest, regression trees and support vector machines. Ore Geology Reviews, 71, 804–818.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (Grant: 81573603), the National Natural Science Foundation of China (Grant: 81303224).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Zhou, M., Du, W., Qin, K. et al. Distinguish Crude and Sweated Chinese Herbal Medicine with Support Vector Machine and Random Forest Methods. Wireless Pers Commun 102, 1827–1838 (2018). https://doi.org/10.1007/s11277-017-5239-3
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11277-017-5239-3