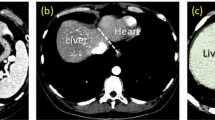
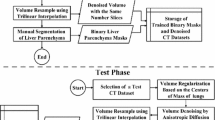
Abstract
The issues based on gastroenterology are the most distressing diseases in a human anatomy with the largest solid organ, liver that requires serious attention in its diagnosis. The challenging task of Liver segmentation is the main motive, proposed with the exploring of level set methodology with signed pressure force combined with masking and other Morphological operations. Next, the segmented liver multi-atlases are condensed together as a complete 3D projection of Human Liver in 360° shape angulation. The huge set of abdominal slices with 0.16 mm spacing of about 250 real time images of each subject, is treated as input with this proposed method and nearly 2550 CT images are used to get segmented using this methodology. The Evaluation Metrics given as 5 Error Metrics and 4 Performance Metrics obtained with the averaging of 10 atlases to each point, of the segmented atlases, outperforms the comparative methods of the Radiologist’s inference of liver boundary detection and an additional existing Local Fuzzy thresholding methodology. The overall visual outcome, evaluation metrical values and graphical data shows the peculiar performance of proposed methodology. This outcome with the electronic evolution of computed tomography appends to the wireless sensory waves to mark the CT images for our input. The doctors will be greatly helped by our process, to serve the global cause of saving lives by diagnosing accurately the complications of liver with appropriate segments, at early stage itself.








Similar content being viewed by others
References
Maton, A., Hopkins, J., Johnson, S., McLaughlin, D. L. C. W., Warner, M. Q., & Wright, J. D. (1993). Human biology and health (Print book.). Englewood Cliffs : Prentice Hall. Retrieved from http://www.worldcat.org/title/human-biology-and-health/oclc/32308337.
Tucker, M. E. (2013). Global burden of liver disease substantial. Medscape Medical News, The Liver Meeting 2013: American Association for the Study of Liver Diseases (AASLD). https://www.medscape.com/viewarticle/813788.
Sheron, N. (2013). Facts-about-liver-disease. Retrieved from https://www.britishlivertrust.org.uk/about-us/media-centre/facts-about-liver-disease/. Accessed 16 Aug 2017.
Campbell, A. (2017). Alcohol-related deaths in the UK: Registered in 2015. Office for National Statistics. https://www.ons.gov.uk/peoplepopulationandcommunity/healthandsocialcare/causesofdeath/bulletins/alcoholrelateddeathsintheunitedkingdom/registeredin2015. Accessed 16 Feb 2017.
Liver cancer statistics. (2012). World cancer research fund international. http://www.wcrf.org/int/cancer-facts-figures/data-specific-cancers/liver-cancer-statistics. Accessed 3 Aug 2017.
World Health Rankings. (2017). WHO 2013. http://www.worldlifeexpectancy.com/cause-of-death/liver-cancer/by-country/. Accessed 12 July 2017.
Chowdhury, A. (2004). Liver diseases in adults. Liver Foundation India. Retrieved from http://www.theliverfoundationindia.org/liver-disease-in-adults.html. Accessed 5 May 2017.
Dvořák, P., Bartušek, K., Kropatsch, W. G., & Smékal, Z. (2015). Automated multi-contrast brain pathological area extraction from 2D MR images. Journal of Applied Research and Technology,13(1), 58–69. https://doi.org/10.1016/S1665-6423(15)30005-5.
Kim, J., Lee, S., Lee, G., Park, Y., & Hong, Y. (2016). Using a method based on a modified k-means clustering and mean shift segmentation to reduce file sizes and detect brain tumors from magnetic resonance (MRI) images. Wireless Personal Communications,89(3), 993–1008. https://doi.org/10.1007/s11277-016-3420-8.
Cootes, T., Hill, A., Taylor, C., & Haslam, J. (1994). Use of active shape models for locating structures in medical images. Image and Vision Computing,12(6), 355–365. https://doi.org/10.1016/0262-8856(94)90060-4.
Berlin, I., Lamecker, H., Lange, T., Seebass, M., & Berlin, I. (2004). Segmentation of the liver using a 3D statistical shape model, April 9. Retrieved from https://opus4.kobv.de/opus4-zib/frontdoor/index/index/docId/784.
Kainmüller, D., Lange, T., & Lamecker, H. (2007). Shape constrained automatic segmentation of the liver based on a heuristic intensity model. In MICCAI workshop on 3D segmentation in the clinic: A grand challenge, (pp. 109–116). http://mbi.dkfz-heidelberg.de/grandchallenge2007/sites/proceed.htm.
Rodriguez, R., Mexicano, A., Bila, J., Cervantes, S., & Ponce, R. (2015). Feature extraction of electrocardiogram signals by applying adaptive threshold and principal component analysis. Journal of Applied Research and Technology,13(2), 261–269. https://doi.org/10.1016/j.jart.2015.06.008.
Zhang, Xing, Tian, Jie, Deng, Kexin, Yongfang, Wu, & Li, Xiuli. (2010). Automatic liver segmentation using a statistical shape model with optimal surface detection. IEEE Transactions on Biomedical Engineering,57(10), 2622–2626. https://doi.org/10.1109/TBME.2010.2056369.
Heimann, T., Meinzer, H., & Wolf, I. (2007). A statistical deformable model for the segmentation of liver CT volumes using extended training data. In Proceeding on MICCAI workshop 3-D segmentat. Clinic: A grand, challenge, (pp. 161–166). https://scholar.google.com/scholar?q=Heimann%2C%20T.%2C%20Meinzer%2C%20H.P.%2C%20Wolf%2C%20I.%3A%20A%20Statistical%20Deformable%20Model%20for%20the%20Segmentation%20of%20Liver%20CT%20Volumes.%20In%3A%203D%20Segmentation%20in%20the%20Clinic%20-%20A%20Grand%20Challenge%2C%20pp.%20161%E2%80%93166%20%282007%290.
Bellman, R. (1966). Dynamic programming. Science,153(3731), 34–37. https://doi.org/10.1126/science.153.3731.34.
Seghers, D., Slagmolen, P., Lambelin, Y., Hermans, J., Loeckx, D., Maes, F., & Suetens, P. (2007). Landmark based liver segmentation using local shape and local intensity models. In Proceedings of workshop of the 10th international conference on MICCAI, workshop on 3D segmentation in the clinic: A grand challenge, (pp. 135–142).
Saddi, K. A., Rousson, M., Chefd’hotel, C., & Cheriet, F. (2007). Global-to-local shape matching for liver segmentation in CT imaging. In Proceedings of MICCAI workshop on 3D segmentation in the clinic: a grand challenge, (pp. 207–214).
Schmidt, G., Athelogou, M., Schönmeyer, R., Korn, R., & Binnig, G. (2007). Cognition network technology for a fully automated 3d segmentation of liver. In MICCAI workshop on 3D segmentation in the clinic: A grand challenge (pp. 125–133). http://mbi.dkfzheidelberg.de/grand-challenge2007/sites/proceed.htm. http://mbi.dkfz-heidelberg.de/grand-challenge2007/web/p125.pdf.
Chi, Y., Cashman, P. M. M., Bello, F., & Kitney, R. I. (2007). A discussion on the evaluation of a new automatic liver volume segmentation method for specified CT image datasets. In: Proceedings of MICCAI workshop on 3D segmentation in the clinic: a grand challenge (pp. 167–175). https://scholar.google.com/scholar?q=Chi%20Y%2C%20Cashman%20PMM%2C%20Bello%20F%2C%20Kitney%20RI%2C%282007%29%20A%20discussion%20on%20the%20evaluation%20of%20a%20new%20automatic%20liver%20volume%20segmentation%20method%20for%20specified%20CT%20image%20datasets.%20In%3A%20Proceedings%20of%20MICCAI%20workshop%20on%203D%20segmentation%20in%20the%20clinic%3A%20a%20grand%20challenge%2C%20pp%20167%E2%80%93175.
Susomboon, R., Raicu, D. S., & Furst, J. (2007). A hybrid approach for liver segmentation. In: Proceedings of MICCAI workshop on 3D segmentation in the clinic: a grand challenge (pp. 151–160). https://scholar.google.com/scholar?q=Susomboon%20R%2C%20Raicu%20DS%2C%20Furst%20J%20%282007%29%20A%20hybrid%20approach%20for%20liver%20segmentation.%20In%3A%20Proceedings%20of%20MICCAI%20workshop%20on%203D%20segmentation%20in%20the%20clinic%3A%20a%20grand%20challenge%2C%20pp%20151%E2%80%93160.
McLachlan, G. J., & Krishnan, T. (2008). The EM algorithm and extensions, 2E. Hoboken: Wiley. https://doi.org/10.1002/9780470191613.
Haralick, R. M., Shanmugam, K., & Dinstein, I. (1973). Textural features for image classification. IEEE Transactions on Systems, Man, and Cybernetics, SMC,3(6), 610–621. https://doi.org/10.1109/tsmc.1973.4309314.
Duda, R. O., Hart, P. E., & Stork, D. G. (2000). Pattern classification (2nd ed.). New York: Wiley.
Beichel, R., Bauer, C., Bornik, A., Sorantin, E., & Bischof, H. (2007). Liver segmentation in CT data : A segmentation refinement approach. In: Proceedings of MICCAI workshop on 3D segmentation in the clinic: a grand Challenge (pp 235–245). https://scholar.google.com/scholar?q=Beichel%20R%2C%20Bauer%20C%2C%20Bornik%20A%2C%20Sorantin%20E%2C%20Bischof%20H%20%282007%29%20Liver%20segmentation%20in%20CT%20data%3A%20a%20segmentation%20refinement%20approach.%20In%3A%20Proceedings%20of%20MICCAI%20workshop%20on%203D%20segmentation%20in%20the%20clinic%3A%20a%20grand%20Challenge%2C%20pp%20235%E2%80%93245.
Boykov, Y., & Funka-Lea, G. (2006). Graph cuts and efficient N-D image segmentation. International Journal of Computer Vision,70(2), 109–131. https://doi.org/10.1007/s11263-006-7934-5.
Beck, A., & Aurich, V. (2007). HepaTux—A semiautomatic liver segmentation system. In: Proceedings of MICCAI workshop on 3Dsegmentation in the clinic: a grand challenge (pp. 225–233). https://scholar.google.com/scholar?q=Beck%20A%2C%20Aurich%20V%20%282007%29%20HepaTuxa%20semiautomatic%20liver%20segmentation%20system.%20In%3A%20Proceedings%20of%20MICCAI%20workshop%20on%203D%20segme ntation%20in%20the%20clinic%3A%20a%20grand%20challenge%2C%20pp%20225%96234.
Rueckert, D., Sonoda, L. I., Hayes, C., Hill, D. L. G., Leach, M. O., & Hawkes, D. J. (1999). Nonrigid registration using free-form deformations: application to breast MR images. IEEE Transactions on Medical Imaging,18(8), 712–721. https://doi.org/10.1109/42.796284.
Slagmolen, P., Elen, A., Seghers, D., Loeckx, D., Maes, F., & Haustermans, K. (2007). Atlas based liver segmentation using nonrigid registration with a B-spline transformation model. In Proceedings of MICCAI workshop on 3D segmentation in the clinic: a grand challenge (pp.197–206). https://scholar.google.com/scholar?q=Slagmolen%20P%2C%20Elen%20A%2C%20Seghers%20D%2C%20Loeckx%20D%2C%20Maes%20F%2C%20Haustermans%2C%20K%20%282007%29%20Atlas%20based%20liver%20segmentation%20using%20nonrigid%20registration%20with%20a%20Bspline%20transformation%20model.%20In%3A%20Proceedings%20of%20MICCAI%20workshop%20on%203D%20segmentation%20in%20the%20clinic%3A%20a%20grand%20challenge%2C%20pp%20197%E2%80%93206.
Sonka, M., Hlavac, V., & Boyle, R. (2007). Image processing, analysis, and machine vision (3rd ed.). Thomson-Engineering.
Adams, R., & Bischof, L. (1994). Seeded region growing. IEEE Transactions on Pattern Analysis and Machine Intelligence,16(6), 641–647. https://doi.org/10.1109/34.295913.
Yuqian Zhao, Yunlong Zan, Xiaofang Wang, & Guiyuan Li. (2010). Fuzzy C-means clustering-based multilayer perceptron neural network for liver CT images automatic segmentation. In 2010 Chinese control and decision conference (pp. 3423–3427). IEEE. https://doi.org/10.1109/ccdc.2010.5498558.
Foruzan, A. H., & Chen, Y.-W. (2013). Segmentation of liver in low-contrast images using k-means clustering and geodesic active contour algorithms. IEICE Transactions on Information and Systems,96(4), 798–807.
Huang, W., Tan, Z. M., Lin, Z., Huang, G., Zhou, J., Chui, C. K., Su, Y. & Chang, S. (2012). A semi-automatic approach to the segmentation of liver parenchyma from 3D CT images with extreme learning machine. In 2012 annual international conference of the IEEE engineering in medicine and biology society (pp. 3752–3755). IEEE. https://doi.org/10.1109/embc.2012.6346783.
Wu, W., Zhou, Z., Wu, S., & Zhang, Y. (2016). Automatic liver segmentation on volumetric CT images using supervoxel-based graph cuts. Computational and Mathematical Methods in Medicine,2016, 1–14. https://doi.org/10.1155/2016/9093721.
Osher, S., & Sethian, J. A. (1988). Fronts propagating with curvature-dependent speed: Algorithms based on Hamilton–Jacobi formulations. Journal of Computational Physics,79(1), 12–49. https://doi.org/10.1016/0021-9991(88)90002-2.
Furukawa, D., Shimizu, A., & Kobatake, H. (2007). Automatic liver segmentation method based on maximum a posterior probability estimation and level set method. In: Proceedings of MICCAI Workshop on 3D Segmentation in the Clinic: A Grand Challenge (pp. 117–124). https://scholar.google.com/scholar?q=Furukawa%2C%20D.%2C%20Shimizu%2C%20A.%2C%20Kobatake%2C%20H.%3A%20Automatic%20liver%20segmentation%20method%20based%20on%20maximum%20a%20posterior%20probability%20estimation%20and%20level%20set%20method.%20In%3A%20Proceedings%20of%20MICCAI%20Workshop%20on%203D%20Segmentation%20in%20the%20Clinic%3A%20A%20Grand%20Challenge%2C%20pp.%20117%E2%80%93124%20%282007%29.
Tomoshige, S., Oost, E., Shimizu, A., Watanabe, H., & Nawano, S. (2014). A conditional statistical shape model with integrated error estimation of the conditions; Application to liver segmentation in non-contrast CT images. Medical Image Analysis,18(1), 130–143. https://doi.org/10.1016/j.media.2013.10.003.
Sethian, J. A. (1999). Level set methods and fast marching methods: Evolving interfaces in computational geometry, fluid mechanics, computer vision and material science. Cambridge University Press.
Malladi, R., Sethian, J. A., & Vemuri, B. C. (1995). Shape modeling with front propagation: A level set approach. IEEE Transactions on Pattern Analysis and Machine Intelligence,17(2), 158–175. https://doi.org/10.1109/34.368173.
Lefohn, A. E., Kniss, J. M., Hansen, C. D., & Whitaker, R. T. (2004). A streaming narrow-band algorithm: interactive computation and visualization of level sets. IEEE Transactions on Visualization and Computer Graphics,10(4), 422–433. https://doi.org/10.1109/TVCG.2004.2.
Caselles, V., Kimmel, R., & Sapiro, G. (1997). Geodesic active contours. International Journal of Computer Vision,22(1), 61–79. https://doi.org/10.1023/A:1007979827043.
Dawant, B. M., Li, R., Lennon, B., & Li, S. (2007). Semi-automatic segmentation of the liver and its evaluation on the MICCAI 2007 grand challenge data set. In: Proceedings of MICCAI workshop on 3D segmentation in the clinic: a grand challenge (pp. 215–221). https://scholar.google.com/scholar?q=Dawant%20BM%2C%20Li%20R%2C%20Lennon%20B%2C%20Li%20S%20%282007%29%20Semiautomatic%20segmentation%20of%20the%20liver%20and%20its%20evaluation%20on%20the%20MICCAI%202007%20grand%20challenge%20data%20set.%20In%3A%20Proceedings%20of%20MICCAI%20workshop%20on%203D%20segmentation%20in%20the%20clinic%3A%20a%20grand%20challenge%2C%20pp%20215%E2%80%93221.
Lee, J., Kim, N., Lee, H., Seo, J. B., & Won, H. J. (2007). Efficient liver segmentation exploiting level-set speed images with 2. Shape Propagation,5D, 189–196.
Wimmer, A., Soza, G., & Hornegger, J. (2007). Two-stage semi-automatic organ segmentation framework using radial basis functions and level sets. In Proceedings of MICCAI workshop on 3D segmentation in the clinic: a grand challenge (pp. 179–188). https://scholar.google.com/scholar?q=Wimmer%20A%2C%20Soza%20G%2C%20Hornegger%20J%20%282007%29%20Two-stage%20semiautomatic%20organ%20segmentation%20framework%20using%20radial%20basis%20functions%20and%20level%20sets%3A%20In%3A%20Proceedings%20of%20MICCAI%20workshop%20on%203D%20segmentation%20in%20the%20clinic%3A%20a%20grand%20challenge%2C%20pp%20179%E2%80%93188.
Masoodhu Banu, N. M., & Sujatha, S. (2015). Improved tampering detection for image authentication based on image partitioning. Wireless Personal Communications,84(1), 69–85. https://doi.org/10.1007/s11277-015-2594-9.
Lee, S., Lee, G., Hong, Y., & Kim, J. (2016). A study on the improved normalized cut algorithm using a bilateral filter for efficient object extraction from image. Wireless Personal Communications,86(1), 77–90. https://doi.org/10.1007/s11277-015-3033-7.
Hongzhe Y., Yongtian W., Jian Y., & Yue L. (2010). A novel graph cuts based liver segmentation method. In 2010 International conference of medical image analysis and clinical application (pp. 50–53). IEEE. https://doi.org/10.1109/miaca.2010.5528409.
Massoptier, L., & Casciaro, S. (2007). Fully automatic liver segmentation through graph-cut technique. In 2007 29th Annual international conference of the IEEE engineering in medicine and biology society (pp. 5243–5246). IEEE. https://doi.org/10.1109/iembs.2007.4353524.
Mittal, A., Kundu, C., Bose, R., & Shevgaonkar, R. K. (2017). Entropy based image segmentation for energy efficient LTE system with cloud. Wireless Personal Communications,92(3), 1145–1162. https://doi.org/10.1007/s11277-016-3598-9.
Luo, S., Li, X., & Li, J. (2014). Review on the methods of automatic liver segmentation from abdominal images. Journal of Computer and Communications,2(2), 1–7. https://doi.org/10.4236/jcc.2014.22001.
Campadelli, P., Casiraghi, E., & Esposito, A. (2009). Liver segmentation from computed tomography scans: A survey and a new algorithm. Artificial Intelligence in Medicine,45(2–3), 185–196. https://doi.org/10.1016/j.artmed.2008.07.020.
Rathore, S., Iftikhar, M. A., Hussain, M., & Jalil, A. (2011). Texture analysis for liver segmentation and classification: A survey. Frontiers of Information Technology,2011, 121–126. https://doi.org/10.1109/FIT.2011.30.
Mharib, A. M., Ramli, A. R., Mashohor, S., & Mahmood, R. B. (2012). Survey on liver CT image segmentation methods. Artificial Intelligence Review,37(2), 83–95. https://doi.org/10.1007/s10462-011-9220-3.
Chan, T. F., & Vese, L. A. (2001). Active contours without edges. IEEE Transactions on Image Processing,10(2), 266–277. https://doi.org/10.1109/83.902291.
Pomerantz, S. M., White, C. S., Krebs, T. L., Daly, B., Sukumar, S. A., Hooper, F., et al. (2000). Liver and bone window settings for soft-copy interpretation of chest and abdominal CT. American Journal of Roentgenology,174(2), 311–314. https://doi.org/10.2214/ajr.174.2.1740311.
Patten, R. M., Gunberg, S. R., Brandenburger, D. K., & Richardson, M. L. (2000). CT detection of hepatic and splenic injuries. American Journal of Roentgenology,175(4), 1107–1110. https://doi.org/10.2214/ajr.175.4.1751107.
Dang, T., & Mandarano, G. (2006). A review of the benefits and rationale of viewing liver window settings for abdominal computed tomography scans. The Radiographer,53(1), 12–19.
Woodward, A., Chan, Y. H., Gong, R., Nguyen, M., Gee, T., Delmas, P., et al. (2017). A low cost framework for real-time marker based 3-D human expression modeling. Journal of Applied Research and Technology,15(1), 61–77. https://doi.org/10.1016/j.jart.2017.01.002.
Yang, S.-C., Yu, C.-Y., Lin, C.-J., Lin, H.-Y., & Lin, C.-Y. (2015). Reconstruction of three-dimensional breast-tumor model using multispectral gradient vector flow snake method. Journal of Applied Research and Technology,13(2), 279–290. https://doi.org/10.1016/j.jart.2015.06.014.
Zhang, R., Zhu, S., & Zhou, Q. (2016). A novel gradient vector flow snake model based on convex function for infrared image segmentation. Sensors,16(10), 1756. https://doi.org/10.3390/s16101756.
Zhu, S., & Gao, R. (2016). A novel generalized gradient vector flow snake model using minimal surface and component-normalized method for medical image segmentation. Biomedical Signal Processing and Control. https://doi.org/10.1016/j.bspc.2015.12.004.
Singh, A. K., Dave, M., & Mohan, A. (2015). Multilevel encrypted text watermarking on medical images using spread-spectrum in DWT domain. Wireless Personal Communications,83(3), 2133–2150. https://doi.org/10.1007/s11277-015-2505-0.
Jiji, G. W., & DuraiRaj, P. J. (2015). Content-based image retrieval techniques for the analysis of dermatological lesions using particle swarm optimization technique. Applied Soft Computing,30, 650–662. https://doi.org/10.1016/j.asoc.2015.01.058.
Gimel, G. (2013). Part 3: Image processing. Basics of Mathematical Morphology. https://www.cs.auckland.ac.nz/courses/compsci373s1c/PatricesLectures/2013/CS373-IP-02.pdf.
Efford, N. (2002). Digital image processing: A practical introduction using JavaTM. Pearson Education. Retrieved from https://www.cs.auckland.ac.nz/courses/compsci773s1c/lectures/ImageProcessing-html/topic4.htm.
Ronse, C., & Devijve, P. A. (1984). Connected components in binary images: The detection problem. Research Studies (pp 91-102). https://scholar.google.com/scholar?as_q=%22Connected+Components+in+Binary+Images%3A+The+Detection+Problem%2C%22&as_occt=title&hl=en&as_sdt=0%2C31.
Yang, X. D. (1989). An improved algorithm for labeling connected components in a binary image. TR 89-981, March. https://apps.dtic.mil/dtic/tr/fulltext/u2/a210100.pdf.
Park, J.-M., Looney, C.G., & Chen, H.-C. (2000) Fast connected component labeling algorithm using a divide and conquer technique. In S.Y. Shin (Ed.), Computers and Their Applications: Proceedings of the ISCA 15th International Conference on Computers and Their Applications, March 29-31, 2000. ISCA 2000 (pp. 373–376). New Orleans: Louisiana USA.
Zhang, K., Zhang, L., Song, H., & Zhou, W. (2010). Active contours with selective local or global segmentation: A new formulation and level set method. Image and Vision Computing,28(4), 668–676. https://doi.org/10.1016/j.imavis.2009.10.009.
Jiang, H., Feng, R., & Gao, X. (2011). Level set based on signed pressure force function and its application in liver image segmentation. Wuhan University Journal of Natural Sciences.. https://doi.org/10.1007/s11859-011-0748-5.
Aja-Fernández, S., Curiale, A. H., & Vegas-Sánchez-Ferrero, G. (2015). A local fuzzy thresholding methodology for multiregion image segmentation. Knowledge-Based Systems,83, 1–12. https://doi.org/10.1016/j.knosys.2015.02.029.
Acknowledgements
The Abdominal CT slices were collected from TVMCH, Medall Diagnostics, Tirunelveli and a few more from Arthi Scans, Tirunelveli. Our special thanks to Dr.Arun Kumar MD.,RD, Managing Director, Arthi Scans-Tirunelveli for rendering his support in marking of referential images.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Rajathi, G.I., Jiji, G.W. A Novel Automatic Liver Segmentation by Level Set Method Over Real-Time Sensory Computed Tomography. Wireless Pers Commun 109, 1987–2010 (2019). https://doi.org/10.1007/s11277-019-06664-9
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11277-019-06664-9