Abstract
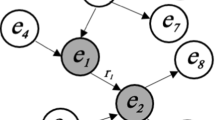
Structured entities analysis is the basis of the modern science, such as chemical science, biological science, environmental science and medical science. Recently, a huge amount of computational models have been proposed to analyze structured entities such as chemical molecules and proteins. However, the problem becomes complex when local structural entity graphs and a global entity interaction graph are both involved. The unique graph of graphs structure cannot be properly exploited by most existing works for structural entity analysis. Some works that build neural networks on the graph of graphs cannot preserve the local graph structure effectively, hence, reducing the expressive power of the model. In this paper, we propose a Powerful Graph Of graphs neural Network, namely PGON, which has 3-Weisfeiler-Lehman expressive power and captures the attributes and structural information from both structured entity graphs and entity interaction graph hierarchically. Extensive experiments are conducted on real-world datasets, which show that PGON outperforms other state-of-the-art methods on both graph classification and graph interaction prediction tasks.






Similar content being viewed by others
References
Borgwardt, K.M., Ong, C.S., Schönauer, S., Vishwanathan, S.V.N., Smola, A.J., Kriegel, H.: Protein function prediction via graph kernels. In: Proceedings Thirteenth International Conference on Intelligent Systems for Molecular Biology 2005, 25-29 June 2005, pp 47–56, Detroit, MI USA (2005)
Cai, J.Y., Fürer, M., Immerman, N.: An optimal lower bound on the number of variables for graph identification. Combinatorica 12(4), 389–410 (1992)
Chu, X., Lin, Y., Wang, Y., Wang, L., Wang, J., Gao, J.: Mlrda: a multi-task semi-supervised learning framework for drug-drug interaction prediction. In: IJCAI (2019)
D’Agostino, G., Scala, A.: Networks of Networks: the Last Frontier of Complexity, vol. 340. Springer, New York (2014)
Debnath, A.K., Lopez de Compadre, R.L., Debnath, G., Shusterman, A.J., Hansch, C.: Structure-activity relationship of mutagenic aromatic and heteroaromatic nitro compounds. correlation with molecular orbital energies and hydrophobicity. J. Med. Chem. 34(2), 786–797 (1991)
Devlin, J., Chang, M.W., Lee, K., Toutanova, K.: Bert: Pre-training of deep bidirectional transformers for language understanding. arXiv:1810.04805 (2018)
Dobson, P.D., Doig, A.J.: Distinguishing enzyme structures from non-enzymes without alignments. J. Mol. Biol. 330(4), 771–783 (2003)
Dong, G., Gao, J., Du, R., Tian, L., Stanley, H.E., Havlin, S.: Robustness of network of networks under targeted attack. Phys. Rev. E 87(5), 052804 (2013)
Errica, F., Podda, M., Bacciu, D., Micheli, A.: A fair comparison of graph neural networks for graph classification. arXiv:1912.09893 (2019)
Fey, M., Lenssen, J.E., Morris, C., Masci, J., Kriege, N.M.: Deep graph matching consensus. In: International Conference on Learning Representations (2019)
Grohe, M.: Descriptive complexity, canonisation, and definable graph structure theory, vol. 47. Cambridge University Press, Cambridge (2017)
Grohe, M., Otto, M.: Pebble games and linear equations. J. Symb. Log. pp. 797–844 (2015)
Hamilton, W., Ying, Z., Leskovec, J.: Inductive representation learning on large graphs. In: NIPS, pp 1024–1034 (2017)
Hao, Y., Cao, X., Fang, Y., Xie, X., Wang, S.: Inductive link prediction for nodes having only attribute information. arXiv:2007.08053 (2020)
Harada, S., Akita, H., Tsubaki, M., Baba, Y., Takigawa, I., Yamanishi, Y., Kashima, H.: Dual graph convolutional neural network for predicting chemical networks. BMC Bioinformatics 21, 1–13 (2020)
Kersting, K., Kriege, N.M., Morris, C., Mutzel, P., Neumann, M.: Benchmark data sets for graph kernels. http://graphkernels.cs.tu-dortmund.de (2016)
Kipf, T.N., Welling, M.: Semi-supervised classification with graph convolutional networks. In: ICLR, p 2017 (2017)
Kwon, S., Yoon, S.: Deepcci: End-to-end deep learning for chemical-chemical interaction prediction. In: Proceedings of the 8th ACM International Conference on Bioinformatics, Computational Biology, and Health Informatics, pp 203–212 (2017)
Landrum, G.: Rdkit documentation. Release 1, 1–79 (2013)
Lee, J., Lee, I., Kang, J.: Self-attention graph pooling. arXiv:1904.08082 (2019)
Lee, J.B., Rossi, R., Kong, X.: Graph classification using structural attention. In: SIGKDD, pp 1666–1674. ACM (2018)
Li, J., Rong, Y., Cheng, H., Meng, H., Huang, W., Huang, J.: Semi-supervised graph classification: A hierarchical graph perspective. In: Liu, L., White, R.W., Mantrach, A., Silvestri, F., McAuley, J.J., Baeza-Yates, R., Zia, L. (eds.) The World Wide Web Conference, WWW 2019, May 13-17, 2019, pp 972–982. ACM, San Francisco, CA, USA (2019)
Li, Y., Gu, C., Dullien, T., Vinyals, O., Kohli, P.: Graph matching networks for learning the similarity of graph structured objects. arXiv:1904.12787 (2019)
Lian, D., Liu, Q., Chen, E.: Personalized ranking with importance sampling. In: WWW, vol. 2020, pp 1093–1103 (2020)
Lian, D., Wang, H., Liu, Z., Lian, J., Chen, E., Xie, X.: Lightrec: A memory and search-efficient recommender system. In: WWW, vol. 2020, pp 695–705 (2020). https://doi.org/10.1145/3366423.3380151
Liao, R., Li, Y., Song, Y., Wang, S., Hamilton, W., Duvenaud, D.K., Urtasun, R., Zemel, R.: Efficient graph generation with graph recurrent attention networks. In: Advances in Neural Information Processing Systems, pp 4255–4265 (2019)
Liu, Z., Chen, C., Li, L., Zhou, J., Li, X., Song, L., Qi, Y.: Geniepath: Graph neural networks with adaptive receptive paths. In: AAAI, vol. 33, pp 4424–4431 (2019)
Maron, H., Ben-Hamu, H., Serviansky, H., Lipman, Y.: Provably powerful graph networks. In: Wallach, H.M., Larochelle, H., Beygelzimer, A., d’Alché-Buc, F., Fox, E.B., Garnett R. (eds.) Advances in Neural Information Processing Systems 32: Annual Conference on Neural Information Processing Systems 2019, NeurIPS 2019, 8-14 December 2019, pp 2153–2164, Vancouver, BC, Canada (2019)
Menche, J., Sharma, A., Kitsak, M., Ghiassian, S.D., Vidal, M., Loscalzo, J., Barabási, A. L.: Uncovering disease-disease relationships through the incomplete interactome. Science 347(6224) (2015)
Meng, L., Zhang, J.: Isonn: Isomorphic neural network for graph representation learning and classification. arXiv:1907.09495 (2019)
Mikolov, T., Sutskever, I., Chen, K., Corrado, G.S., Dean, J.: Distributed representations of words and phrases and their compositionality. In: Advances in Neural Information Processing Systems, pp 3111–3119 (2013)
Nguyen, T.D., Phung, D, et al: Unsupervised universal self-attention network for graph classification (2019)
Ni, J., Tong, H., Fan, W., Zhang, X.: Inside the atoms: ranking on a network of networks. In: SIGKDD, pp 1356–1365 (2014)
Rome, E., Langeslag, P., Usov, A.: Federated modelling and simulation for critical infrastructure protection. In: Networks of Networks: The Last Frontier of Complexity, pp 225–253. Springer (2014)
Ryu, J.Y., Kim, H.U., Lee, S.Y.: Deep learning improves prediction of drug–drug and drug–food interactions. Proc. Natl. Acad. Sci. 115(18), E4304–E4311 (2018)
Schlichtkrull, M., Kipf, T.N., Bloem, P., Van Den Berg, R., Titov, I., Welling, M.: Modeling relational data with graph convolutional networks. In: European Semantic Web Conference, pp 593–607. Springer (2018)
Shi, H., Fan, H., Kwok, J.T.: Effective decoding in graph auto-encoder using triadic closure. In: Proceedings of the AAAI Conference on Artificial Intelligence, vol. 34, pp 906–913 (2020)
Veličković, P., Cucurull, G., Casanova, A., Romero, A., Lio, P., Bengio, Y.: Graph attention networks. arXiv:1710.10903 (2017)
Wang, H., Lian, D., Zhang, Y., Qin, L., He, X., Lin, Y., Lin, X.: Binarized graph neural network. World Wide Web, pp. 1–24 (2021)
Wang, H., Lian, D., Zhang, Y., Qin, L., Lin, X.: Gognn: Graph of graphs neural network for predicting structured entity interactions. arXiv:2005.05537 (2020)
Weininger, D., Weininger, A., Weininger, J.L.: Smiles. 2. algorithm for generation of unique smiles notation. J. Chem. Inf. Comput. Sci. 29(2), 97–101 (1989)
Xu, K., Hu, W., Leskovec, J., Jegelka, S.: How powerful are graph neural networks?. In: 7th International Conference on Learning Representations, ICLR 2019, May 6-9, 2019. OpenReview.net, New Orleans, LA, USA (2019)
Xu, N., Wang, P., Chen, L., Tao, J., Zhao, J.: Mr-gnn: Multi-resolution and dual graph neural network for predicting structured entity interactions. arXiv:1905.09558 (2019)
Yang, C., Zhuang, P., Shi, W., Luu, A., Li, P.: Conditional structure generation through graph variational generative adversarial nets. In: Advances in Neural Information Processing Systems, pp 1340–1351 (2019)
You, J., Liu, B., Ying, Z., Pande, V., Leskovec, J.: Graph convolutional policy network for goal-directed molecular graph generation. In: Advances in neural information processing systems, pp 6410–6421 (2018)
Zhang, M., Chen, Y.: Link prediction based on graph neural networks. In: NIPS, pp 5165–5175 (2018)
Zhang, M., Cui, Z., Neumann, M., Chen, Y.: An end-to-end deep learning architecture for graph classification. In: AAAI (2018)
Zhang, Z., Bu, J., Ester, M., Zhang, J., Yao, C., Yu, Z., Wang, C.: Hierarchical graph pooling with structure learning. arXiv:1911.05954(2019)
Zitnik, M., Agrawal, M., Leskovec, J.: Modeling polypharmacy side effects with graph convolutional networks. Bioinformatics 34(13), i457–i466 (2018)
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article belongs to the Topical Collection: Special Issue on Large Scale Graph Data Analytics Guest Editors: Xuemin Lin, Lu Qin, Wenjie Zhang, and Ying Zhang
Rights and permissions
About this article
Cite this article
Wang, H., Lian, D., Liu, W. et al. Powerful graph of graphs neural network for structured entity analysis. World Wide Web 25, 609–629 (2022). https://doi.org/10.1007/s11280-021-00900-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11280-021-00900-8