Abstract
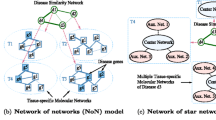
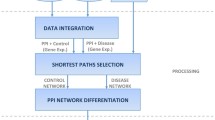
Prioritization of candidate disease genes is crucial for improving medical care, and is one of the fundamental challenges in the post-genomic era. In recent years, different network-based methods for gene prioritization are proposed. Previous studies on gene prioritization show that tissue-specific protein-protein interaction (PPI) networks built by integrating PPIs with tissue-specific gene expression profiles can perform better than tissue-na¨ıve global PPI network. Based on the observations that diseases with similar phenotypes are likely to have common related genes, and genes associated with the same phenotype tend to interact with each other, we propose a method to prioritize disease genes based on a heterogeneous network built by integrating phenotypic features and tissue-specific information. In this heterogeneous network, the PPI network is built by integrating phenotypic features with a tissue-specific PPI network, and the disease network consists of the diseases that are associated with the same phenotype and tissue as the query disease. To determine the impacts of these two factors on gene prioritization, we test three typical network-based prioritization methods on heterogeneous networks consisting of combinations of different PPIs and disease networks built with or without phenotypic features and tissue-specific information. We also compare the proposed method with other tissuespecific networks. The results of case studies reveals that integrating phenotypic features with a tissue-specific PPI network improves the prioritization results. Moreover, the disease networks generated using our method not only show comparable performance with the widely used disease similarity dataset of 5080 human diseases, but are also effective for diseases that are not in the dataset.
Similar content being viewed by others
References
Ritchie M D, Holzinger E R, Li R, et al. Methods of integrating data to uncover genotype-phenotype interactions. Nat Rev Genet, 2015, 16: 85–97
Moreau Y, Tranchevent L-C. Computational tools for prioritizing candidate genes: boosting disease gene discovery. Nat Rev Genet, 2012, 13: 523–536
Piro R M, Di Cunto F. Computational approaches to disease-gene prediction: rationale, classification and successes. FEBS J, 2012, 279: 678–696
Wang X J, Gulbahce N, Yu H Y. Network-based methods for human disease gene prediction. Brief Funct Genomics, 2011, 10: 280–293
Lan W, Wang J X, Li M, et al. Computational approaches for prioritizing candidate disease genes based on PPI networks. Tsinghua Sci Technol, 2015, 20: 500–512
Wu X B, Jiang R, Zhang M, et al. Network-based global inference of human disease genes. Mol Syst Biol, 2008, 4: 189
Vanunu O, Magger O, Ruppin E, et al. Associating genes and protein complexes with disease via network propagation. PLoS Comput Biol, 2010, 6: e1000641
Li Y J, Patra J C. Genome-wide inferring gene-phenotype relationship by walking on the heterogeneous network. Bioinformatics, 2010, 26: 1219–1224
Wang J X, Peng X Q, Peng W, et al. Dynamic protein interaction network construction and applications. Proteomics, 2014, 14: 338–352
Gaulton K J, Mohlke K L, Vision T J. A computational system to select candidate genes for complex human traits. Bioinformatics, 2007, 23: 1132–1140
Schlicker A, Lengauer T, Albrecht M. Improving disease gene prioritization using the semantic similarity of Gene Ontology terms. Bioinformatics, 2010, 26: i561–i567
Linghu B, Snitkin E S, Hu Z, et al. Genome-wide prioritization of disease genes and identification of disease-disease associations from an integrated human functional linkage network. Genome Biol, 2009, 10: R91
Franke L, van Bakel H, Fokkens L, et al. Reconstruction of a functional human gene network, with an application for prioritizing positional candidate genes. Amer J Hum Genet, 2006, 78: 1011–1025
Robinson P N, Webber C. Phenotype ontologies and cross-species analysis for translational research. PLoS Genet, 2014, 10: e1004268
Hwang S, Kim E, Yang S, et al. MORPHIN: a web tool for human disease research by projecting model organism biology onto a human integrated gene network. Nucl Acids Res, 2014, 42: W147–W153
Winter E E, Goodstadt L, Ponting C P. Elevated rates of protein secretion, evolution, and disease among tissue-specific genes. Genome Res, 2004, 14: 54–61
Chao E C, Lipkin S M. Molecular models for the tissue specificity of DNA mismatch repair-deficient carcinogenesis. Nucl Acids Res, 2006, 34: 840–852
Magger O, Waldman Y Y, Ruppin E, et al. Enhancing the prioritization of disease-causing genes through tissue specific protein interaction networks. PLoS Comput Biol, 2012, 8: e1002690
Prasad T S K, Goel R, Kandasamy K, et al. Human protein reference database2009 update. Nucl Acids Res, 2009, 37: D767–D772
Barshir R, Basha O, Eluk A, et al. The tissuenet database of human tissue protein-protein interactions. Nucl Acids Res, 2013, 41: D841–D844
Su A I, Wiltshire T, Batalov S, et al. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc Nat Acad Sci USA, 2004, 101: 6062–6067
Berglund L, Björling E, Oksvold P, et al. A genecentric human protein atlas for expression profiles based on antibodies. Mol Cell Proteom, 2008, 7: 2019–2027
Bradley R K, Merkin J, Lambert N J, et al. Alternative splicing of RNA triplets is often regulated and accelerates proteome evolution. PLoS Biol, 2012, 10: e1001229
Chatr-aryamontri A, Breitkreutz B-J, Oughtred R, et al. The BioGRID interaction database: 2015 update. Nucl Acids Res, 2015, 43: D470–D478
Salwinski L, Miller C S, Smith A J, et al. The database of interacting proteins: 2004 update. Nucl Acids Res, 2004, 32: D449–D451
Orchard S, Ammari M, Aranda B, et al. The MIntAct project—IntAct as a common curation platform for 11 molecular interaction databases. Nucl Acids Res, 2014, 42: D358–D363
Licata L, Briganti L, Peluso D, et al. MINT, the molecular interaction database: 2012 update. Nucl Acids Res, 2012, 40: D857–D861
Barshir R, Shwartz O, Smoly I Y, et al. Comparative analysis of human tissue interactomes reveals factors leading to tissue-specific manifestation of hereditary diseases. PLoS Comput Biol, 2014, 10: e1003632
Greene C S, Krishnan A, Wong A K, et al. Understanding multicellular function and disease with human tissue-specific networks. Nat Genet, 2015, 47: 569–576
Li M, Zhang J Y, Liu Q, et al. Prediction of disease-related genes based on weighted tissue-specific networks by using DNA methylation. BMC Med Genomics, 2014, 7: S4
Ganegoda G U, Wang J X, Wu F-X, et al. Prediction of disease genes using tissue-specified gene-gene network. BMC Syst Biol, 2014, 8: S3
Jacquemin T, Jiang R. Walking on a tissue-specific disease-protein-complex heterogeneous network for the discovery of disease-related protein complexes. BioMed Res Int, 2013, 2013: 455–458
Robinson P, Krawitz P, Mundlos S. Strategies for exome and genome sequence data analysis in disease-gene discovery projects. Clin Genet, 2011, 80: 127–132
Köhler S, Bauer S, Horn D, et al. Walking the interactome for prioritization of candidate disease genes. Amer J Hum Genet, 2008, 82: 949–958
van Driel M A, Bruggeman J, Vriend G, et al. A text-mining analysis of the human phenome. Eur J Hum Genet, 2006, 14: 535–542
Brunner H G, van Driel M A. From syndrome families to functional genomics. Nat Rev Genet, 2004, 5: 545–551
Yang H, Robinson P N, Wang K. Phenolyzer: phenotype-based prioritization of candidate genes for human diseases. Nat Methods, 2015, 12: 841–843
Javed A, Agrawal S, Ng P C. Phen-Gen: combining phenotype and genotype to analyze rare disorders. Nat Methods, 2014, 11: 935–937
Chen Y, Jiang T, Jiang R. Uncover disease genes by maximizing information flow in the phenome-interactome network. Bioinformatics, 2011, 27: i167–i176
Xie M Q, Hwang T, Kuang R. Prioritizing disease genes by bi-random walk. In: Proceedings of 16th Pacific-Asia Conference on Advances in Knowledge Discovery and Data Mining, Kuala Lumpur, 2012. 292–303
Hamosh A, Scott A F, Amberger J S, et al. Online mendelian inheritance in man (OMIM), a knowledgebase of human genes and genetic disorders. Nucl Acids Res, 2005, 33: D514–D517
Lage K, Hansen N T, Karlberg E O, et al. A large-scale analysis of tissue-specific pathology and gene expression of human disease genes and complexes. Proc Nat Acad Sci, 2008, 105: 20870–20875
Basha O, Flom D, Barshir R, et al. MyProteinNet: build up-to-date protein interaction networks for organisms, tissues and user-defined contexts. Nucl Acids Res, 2015, 43: W258–W263
Köhler S, Doelken S C, Mungall C J, et al. The human phenotype ontology project: linking molecular biology and disease through phenotype data. Nucl Acids Res, 2014, 42: D966–D974
Resnik P. Using information content to evaluate semantic similarity in a taxonomy. In: Proceedings of the 14th International Joint Conference on Artificial Intelligence. San Francisco: Morgan Kaufmann Publishers Inc., 1995. 448–453
Schlicker A, Domingues F, Rahnenf¨uhrer J, et al. A new measure for functional similarity of gene products based on Gene Ontology. BMC Bioinform, 2006, 7: 302
Guo X L, Gao L, Wei C S, et al. A computational method based on the integration of heterogeneous networks for predicting disease-gene associations. PLoS ONE, 2011, 6: e24171
Zhou X Z, Menche J, Barabási A-L, et al. Human symptoms-disease network. Nat Commun, 2014, 5: 4212
Goh K-I, Cusick M E, Valle D, et al. The human disease network. Proc Nat Acad Sci, 2007, 104: 8685–8690
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Deng, Y., Gao, L., Guo, X. et al. Integrating phenotypic features and tissue-specific information to prioritize disease genes. Sci. China Inf. Sci. 59, 070101 (2016). https://doi.org/10.1007/s11432-016-5584-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11432-016-5584-y