Abstract
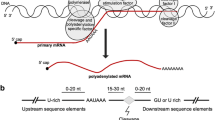
The accurate identification of potential poly(A) sites has contributed to all many studies with regard to alternative polyadenylation. The aim of this study was the development of a machine-learning methodology that will help to discriminate real polyadenylation signals from randomly occurring signals in genomic sequence. Since previous studies have revealed that RNA secondary structure in certain genes has significant impact, the authors tried to computationally pinpoint common structural patterns around the poly(A) sites and to investigate how RNA secondary structure may influence polyadenylation. This involved an initial study on the impact of RNA structure and it was found using motif search tools that hairpin structures might be important. Thus, it was propose that, in addition to the sequence pattern around poly(A) sites, there exists a widespread structural pattern that is also employed during human mRNA polyadenylation. In this study, the authors present a computational model that uses support vector machines to predict human poly(A) sites. The results show that this predictive model has a comparable performance to the current prediction tool. In addition, it was identified common structural patterns associated with polyadenylation using several motif finding programs and this provides new insight into the role of RNA secondary structure plays in polyadenylation.




Similar content being viewed by others
References
Arhin GK et al (2002) Downstream sequence elements with different affinities for the hnRNP H/H’ protein influence the processing efficiency of mammalian polyadenylation signals. Nucleic Acids Res 30(8):1842–1850
Beaudoing E et al (2000) Patterns of variant polyadenylation signal usage in human genes. Genome Res 10(7):1001–1010
Bennett CL et al (2001) A rare polyadenylation signal mutation of the FOXP3 gene (AAUAAA– > AAUGAA) leads to the IPEX syndrome. Immunogenetics 53(6):435–439
Brockman JM et al (2005) PACdb: polya cleavage site and 3′-UTR database. Bioinformatics 21(18):3691–3693
Brown PH, Tiley LS, Cullen BR (1991) Efficient polyadenylation within the human immunodeficiency virus type 1 long terminal repeat requires flanking U3-specific sequences. J Virol 65(6):3340–3343
Carswell S, Alwine JC (1989) Efficiency of utilization of the simian virus 40 late polyadenylation site: effects of upstream sequences. Mol Cell Biol 9(10):4248–4258
Chen CY, Shyu AB (1995) AU-rich elements: characterization and importance in mRNA degradation. Trends Biochem Sci 20(11):465–470
Cheng Y, Miura RM, Tian B (2006) Prediction of mRNA polyadenylation sites by support vector machine. Bioinformatics 22(19):2320–2325
Colgan DF, Manley JL (1997) Mechanism and regulation of mRNA polyadenylation. Genes Dev 11(21):2755–2766
Ding Y, Chan CY, Lawrence CE (2004) Sfold web server for statistical folding and rational design of nucleic acids. Nucleic Acids Res 32(Web Server issue):W135–W141
Gehring NH et al (2001) Increased efficiency of mRNA 3′ end formation: a new genetic mechanism contributing to hereditary thrombophilia. Nat Genet 28(4):389–392
Graber JH et al (1999) In silico detection of control signals: mRNA 3′-end-processing sequences in diverse species. Proc Natl Acad Sci USA 96(24):14055–14060
Hall-Pogar T et al (2005) Alternative polyadenylation of cyclooxygenase-2. Nucleic Acids Res 33(8):2565–2579
Hofacker IL (2003) Vienna RNA secondary structure server. Nucleic Acids Res 31(13):3429–3431
Lee JY et al (2007) PolyA_DB 2: mRNA polyadenylation sites in vertebrate genes. Nucleic Acids Res 35(Database issue):D165–D168
Legendre M, Gautheret D (2003) Sequence determinants in human polyadenylation site selection. BMC Genomics 4(1):7
Liu H et al (2003) An in-silico method for prediction of polyadenylation signals in human sequences. Genome Inform 14:84–93
MacDonald CC, Redondo JL (2002) Reexamining the polyadenylation signal: were we wrong about AAUAAA? Mol Cell Endocrinol 190(1–2):1–8
Macke TJ et al (2001) RNAMotif, an RNA secondary structure definition and search algorithm. Nucleic Acids Res 29(22):4724–4735
Mignone F et al (2005) UTRdb and UTRsite: a collection of sequences and regulatory motifs of the untranslated regions of eukaryotic mRNAs. Nucleic Acids Res 33(Database issue):D141–D146
Moreira A et al (1995) Upstream sequence elements enhance poly(A) site efficiency of the C2 complement gene and are phylogenetically conserved. EMBO J 14(15):3809–3819
Natalizio BJ et al (2002) Upstream elements present in the 3′-untranslated region of collagen genes influence the processing efficiency of overlapping polyadenylation signals. J Biol Chem 277(45):42733–42740
Pruitt KD, Maglott DR (2001) RefSeq and locuslink: NCBI gene-centered resources. Nucleic Acids Res 29(1):137–140
Shaw G, Kamen R (1986) A conserved AU sequence from the 3′ untranslated region of GM-CSF mRNA mediates selective mRNA degradation. Cell 46(5):659–667
Tabaska JE, Zhang MQ (1999) Detection of polyadenylation signals in human DNA sequences. Gene 231(1–2):77–86
Tian B et al (2005) A large-scale analysis of mRNA polyadenylation of human and mouse genes. Nucleic Acids Res 33(1):201–212
Valsamakis A et al (1991) The human immunodeficiency virus type 1 polyadenylylation signal: a 3’ long terminal repeat element upstream of the AAUAAA necessary for efficient polyadenylylation. Proc Natl Acad Sci USA 88(6):2108–2112
Wahle E (1995) 3′-end cleavage and polyadenylation of mRNA precursors. Biochim Biophys Acta 1261(2):183–194
Yan J, Marr TG (2005) Computational analysis of 3′-ends of ESTs shows four classes of alternative polyadenylation in human mouse, and rat. Genome Res 15(3):369–375
Yeo G et al (2004) Variation in alternative splicing across human tissues. Genome Biol 5(10):R74
Zarudnaya MI et al (2003) Downstream elements of mammalian pre-mRNA polyadenylation signals: primary, secondary and higher-order structures. Nucleic Acids Res 31(5):1375–1386
Zhang MQ (2000) Discriminant analysis and its application in DNA sequence motif recognition. Brief Bioinform 1(4):331–342
Zhang XH et al (2003) Sequence information for the splicing of human pre-mRNA identified by support vector machine classification. Genome Res 13(12):2637–2650
Zien A et al (2000) Engineering support vector machine kernels that recognize translation initiation sites. Bioinformatics 16(9):799–807
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Chang, TH., Wu, LC., Chen, YT. et al. Characterization and prediction of mRNA polyadenylation sites in human genes. Med Biol Eng Comput 49, 463–472 (2011). https://doi.org/10.1007/s11517-011-0732-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11517-011-0732-4