Abstract
Glioblastoma multiforme (GBM) are malignant brain tumors, associated with poor overall survival (OS). This study aims to predict OS of GBM patients (in days) using a regression framework and assess the impact of tumor shape features on OS prediction. Multi-channel MR image derived texture features, tumor shape, and volumetric features, and patient age were obtained for 163 GBM patients. In order to assess the impact of tumor shape features on OS prediction, two feature sets, with and without tumor shape features, were created. For the feature set with tumor shape features, the mean prediction error (MPE) was 14.6 days and its 95% confidence interval (CI) was 195.8 days. For the feature set excluding shape features, the MPE was 17.1 days and its 95% CI was observed to be 212.7 days. The coefficient of determination (R2) value obtained for the feature set with shape features was 0.92, while it was 0.90 for the feature set excluding shape features. Although marginal, inclusion of shape features improves OS prediction in GBM patients. The proposed OS prediction method using regression provides good accuracy and overcomes the limitations of GBM OS classification, like choosing data-derived or pre-decided thresholds to define the OS groups.

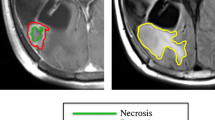
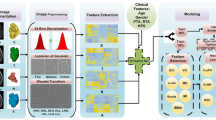
Two feature sets: with and without tumor shape features were extracted from T1-weighted contrast-enhanced, T2-weighted and FLAIR MRI. These feature sets were analyzed using the Mean Prediction Error (MPE) and its 95% Confidence Interval (CI) obtained from the Bland-Altman plot, along with the coefficient of determination (R2) value to assess the impact of tumor shape features on overall survival prediction of glioblastoma multiforme patients.






Similar content being viewed by others
References
Lacroix M, Abi-Said D, Fourney DR, Gokaslan ZL, Shi W, DeMonte F, Lang FF, McCutcheon IE, Hassenbusch SJ, Holland E, Hess K, Michael C, Miller D, Sawaya R (2001) A multivariate analysis of 416 patients with glioblastoma multiforme: prognosis, extent of resection, and survival. J Neurosurg 95:190–198
Hakin-Smith V, Jellinek DA, Levy D, Carroll T, Teo M, Timperley WR, McKay M, Reddel RR, Royds JA (2003) Alternative lengthening of telomeres and survival in patients with glioblastoma multiforme. Lancet 361:836–838
Johnson DR, O’Neill BP (2012) Glioblastoma survival in the United States before and during the temozolomide era. J Neuro-Oncol 107:359–364
Czarnek N, Clark K, Peters KB, Mazurowski MA (2017) Algorithmic three-dimensional analysis of tumor shape in MRI improves prognosis of survival in glioblastoma: a multi-institutional study. J Neuro-Oncol 132:55–62
Drabycz S, Roldán G, De Robles P et al (2010) An analysis of image texture, tumor location, and MGMT promoter methylation in glioblastoma using magnetic resonance imaging. Neuroimage 49:1398–1405
Lee J, Jain R, Khalil K, Griffith B, Bosca R, Rao G, Rao A (2016) Texture feature ratios from relative CBV maps of perfusion MRI are associated with patient survival in glioblastoma. AJNR Am J Neuroradiol 37:37–43
Yang D, Rao G, Martinez J, Veeraraghavan A, Rao A (2015) Evaluation of tumor-derived MRI-texture features for discrimination of molecular subtypes and prediction of 12-month survival status in glioblastoma. Med Phys 42:6725–6735
Zhou M, Chaudhury B, Hall LO, Goldgof DB, Gillies RJ, Gatenby RA (2017) Identifying spatial imaging biomarkers of glioblastoma multiforme for survival group prediction. J Magn Reson Imaging 46:115–123
Macyszyn L, Akbari H, Pisapia JM et al (2015) Imaging patterns predict patient survival and molecular subtype in glioblastoma via machine learning techniques. Neuro-oncology 18:417–425
Bakas S, Akbari H, Sotiras A et al (2017) Advancing the cancer genome atlas glioma MRI collections with expert segmentation labels and radiomic features. Scientific data 4:170117
Bakas S, Reyes M, Jakab A, et al (2018) Identifying the best machine learning algorithms for brain tumor segmentation, progression assessment, and overall survival prediction in the BRATS challenge. arXiv preprint arXiv:1811.02629
Bakas S, Akbari H, Sotiras A et al (2017) Segmentation labels for the pre-operative scans of the TCGA-GBM collection. The Cancer Imaging Archive. https://doi.org/10.7937/K9/TCIA.2017.KLXWJJ1Q
Bakas S, Akbari H, Sotiras A, et al (2017) Segmentation labels for the pre-operative scans of the TCGA-LGG collection. The cancer imaging archive
Menze BH, Jakab A, Bauer S, Kalpathy-Cramer J, Farahani K, Kirby J, Burren Y, Porz N, Slotboom J, Wiest R, Lanczi L, Gerstner E, Weber MA, Arbel T, Avants BB, Ayache N, Buendia P, Collins DL, Cordier N, Corso JJ, Criminisi A, Das T, Delingette H, Demiralp C, Durst CR, Dojat M, Doyle S, Festa J, Forbes F, Geremia E, Glocker B, Golland P, Guo X, Hamamci A, Iftekharuddin KM, Jena R, John NM, Konukoglu E, Lashkari D, Mariz JA, Meier R, Pereira S, Precup D, Price SJ, Raviv TR, Reza SMS, Ryan M, Sarikaya D, Schwartz L, Shin HC, Shotton J, Silva CA, Sousa N, Subbanna NK, Szekely G, Taylor TJ, Thomas OM, Tustison NJ, Unal G, Vasseur F, Wintermark M, Ye DH, Zhao L, Zhao B, Zikic D, Prastawa M, Reyes M, van Leemput K (2015) The multimodal brain tumor image segmentation benchmark (BRATS). IEEE Trans Med Imaging 34:1993–2024
Moshtagh N (2005) Minimum volume enclosing ellipsoid. Convex Optimization 111:112
Bektas S (2015) Least squares fitting of ellipsoid using orthogonal distances. Boletim de Ciências Geodésicas 21:329–339
van Griethuysen JJ, Fedorov A, Parmar C et al (2017) Computational radiomics system to decode the radiographic phenotype. Cancer Res 77:e104–e107
Georgiou H, Mavroforakis M, Dimitropoulos N, Cavouras D, Theodoridis S (2007) Multi-scaled morphological features for the characterization of mammographic masses using statistical classification schemes. Artif Intell Med 41:39–55
Haralick RM, Shanmugam K (1973) Textural features for image classification. IEEE Transactions on systems, man, and cybernetics SMC-3:610–621
Coelho LP (2012) Mahotas: open source software for scriptable computer vision. arXiv preprint arXiv:1211.4907
Van der Walt S, Schönberger JL, Nunez-Iglesias J et al (2014) Scikit-image: image processing in Python. PeerJ 2:e453
Guyon I, Weston J, Barnhill S, Vapnik V (2002) Gene selection for cancer classification using support vector machines. Mach Learn 46:389–422
Hammoud MA, Sawaya R, Shi W, Thall PF, Leeds NE (1996) Prognostic significance of preoperative MRI scans in glioblastoma multiforme. J Neuro-Oncol 27:65–73
Mazurowski MA, Zhang J, Peters KB, Hobbs H (2014) Computer-extracted MR imaging features are associated with survival in glioblastoma patients. J Neuro-Oncol 120:483–488
Mazurowski MA, Czarnek NM, Collins LM, Peters KB, Clark K (2016) Predicting outcomes in glioblastoma patients using computerized analysis of tumor shape: preliminary data. SPIE Proc 9785:97852T–97852T
Sanghani P, Ang BT, King NK, Ren H (2018) Overall survival prediction in glioblastoma multiforme patients from volumetric, shape and texture features using machine learning. Surg Oncol 27(4):709–714
Acknowledgments
This work is supported by the NMRC Bedside Bench under grant R-397-000-245-511 awarded to Dr. Hongliang Ren. This research is also supported by the Singapore Ministry of Health’s National Medical Research Council under its Translational and Clinical Research Flagship Program-Tier 1 (Project No: NMRC/ TCR/ 016-NNI/ 2016).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Sanghani, P., Ang, B.T., King, N.K.K. et al. Regression based overall survival prediction of glioblastoma multiforme patients using a single discovery cohort of multi-institutional multi-channel MR images. Med Biol Eng Comput 57, 1683–1691 (2019). https://doi.org/10.1007/s11517-019-01986-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11517-019-01986-z