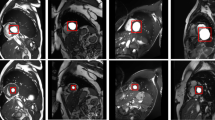
Abstract
Accurate segmentation of the right ventricle (RV) from cardiac magnetic resonance imaging (MRI) images is an essential step in estimating clinical indices such as stroke volume and ejection fraction. Recently, image segmentation methods based on fully convolutional neural networks (FCN) have drawn much attention and shown promising results. In this paper, a new fully automatic RV segmentation method combining the FC-DenseNet and the level set method (FCDL) is proposed. The FC-DenseNet is efficiently trained end-to-end, using RV images and ground truth masks to make a per-pixel semantic inference. As a result, probability images are produced, followed by the level set method responsible for smoothing and converging contours to improve accuracy. It is noted that the iteration times of the level set method is only 4 times, which is due to the semantic segmentation of the FC-DenseNet for RV. Finally, multi-object detection algorithm is applied to locate the RV. Experimental results (including 45 cases, 15 cases for training, 30 cases for testing) show that the FCDL method outperforms the U-net + level set (UL) and the level set methods that use the same dataset and the cardiac functional parameters are computed robustly by the FCDL method. The results validate the FCDL method as an efficient and satisfactory approach to RV segmentation.
Graphical abstract











Similar content being viewed by others
References
Benjamin EJ, Blaha MJ, Chiuve SE, Cushman M, Das SR, Deo R, de Ferranti SD, Floyd J, Fornage M, Gillespie C (2017) Heart disease and stroke statistics-2017 update: a report from the American Heart Association. Circulation 131:e29. https://doi.org/10.1161/CIR.0000000000000491
Roth GA, Johnson C, Abajobir A, Abd-Allah F, Abera SF, Abyu G, Ahmed M, Aksut B, Alam T, Alam K (2017) Global, regional, and national burden of cardiovascular diseases for 10 causes, 1990 to 2015. J Am Coll Cardiol 70:1–25. https://doi.org/10.1016/j.jacc.2017.04.052
Caudron J, Fares J, Lefebvre V, Vivier P-H, Petitjean C, Dacher JN (2012) Cardiac MRI assessment of right ventricular function in acquired heart disease: factors of variability. Acad Radiol 19:991–1002. https://doi.org/10.1016/j.acra.2012.03.022
Luijnenburg SE, Robbers-Visser D, Moelker A, Vliegen HW, Mulder BJM, Helbing WA (2010) Intra-observer and interobserver variability of biventricular function, volumes and mass in patients with congenital heart disease measured by CMR imaging. Int J Card Imaging 26:57–64. https://doi.org/10.1007/s10554-009-9501-y
Mooij CF, de Wit CJ, Graham DA, Powell AJ, Geva T (2008) Reproducibility of MRI measurements of right ventricular size and function in patients with normal and dilated ventricles. J Magn Reson Imaging 28:67–73. https://doi.org/10.1002/jmri.21407
Haddad F, Hunt SA, Rosenthal DN, Murphy DJ (2008) Right ventricular function in cardiovascular disease part I: anatomy physiology aging and functional assessment of the right ventricle. Circulation 117:1436–1448. https://doi.org/10.1161/CIRCULATIONAHA.107.653584
Bristow MR et al (1998) The pressure-overloaded right ventricle in pulmonary hypertension. Chest 114:101S–106S. https://doi.org/10.1378/chest.114.1_supplement.101 s
Ringenberg J, Deo M, Devabhaktuni V et al (2014) Fast, accurate, and fully automatic segmentation of the right ventricle in short-axis cardiac MRI. Comput Med Imaging Graph 38:190–201. https://doi.org/10.1016/j.compmedimag.2013.12.011
Guo Z, Tan W, Wang L et al (2018) Local motion intensity clustering (LMIC) model for segmentation of right ventricle in cardiac MRI images. IEEE J Biomed Health 23:723–730. https://doi.org/10.1109/JBHI.2018.2821709
Ghelich Oghli M, Mohammadzadeh A, Kafieh R et al (2018) A hybrid graph-based approach for right ventricle segmentation in cardiac MRI by long axis information transition. Physica Medica 54:103–116. https://doi.org/10.1016/j.ejmp.2018.09.011
Goshtasby A, Turner DA (1995) Segmentation of cardiac cine MR images for extraction of right and left ventricular chambers. IEEE Trans Med Imaging 14:56–64. https://doi.org/10.1109/42.370402
Liu N, Strugnell W, Slaughter R, Riley R, Crozier S, Wilson S et al (2005) Right ventricle extraction by low level and model-based algorithm. Int Conf Eng Med Biol Soc 2005:1607–1610. https://doi.org/10.1109/IEMBS.2005.1616745
Rosado-Toro JA, Abidov A, Altbach MI et al (2017) Segmentation of the right ventricle in four chamber cine cardiac MR images using polar dynamic programming. Comput Med Imaging Graph 62:15–25. https://doi.org/10.1016/j.compmedimag.2017.08.002
Lötjönen J, Kivistö S, Koikkalainen J, Smutek D, Lauerma K (2004) Statistical shape model of atria, ventricles and epicardium from short- and long-axis MR images. Med Image Anal 8:371–386. https://doi.org/10.1016/J.MEDIA.2004.06.013
ElBaz MS, Fahmy AS (2012) Active shape model with inter-profile modelling paradigm for cardiac right ventricle segmentation. Medical Image Computing and Computer Assisted Interventions (MICCAI), Nice, France, October 2012:691–698
Grosgeorge D, Petitjean C, Caudron J, Fares J, Dacher J-N (2011) Automatic cardiac ventricle segmentation in MR images: a validation study. Int J Comput Assist Radiol Surg 6:573–581. https://doi.org/10.1007/s11548-010-0532-6
Zuluaga MA, Cardoso MJ, Modat M, Ourselin S (2013) Multi-atlas propagation whole heart segmentation from MRI and CTA using a local normalised correlation coefficient criterion. In: International Conference on Functional Imaging & Modeling of the Heart. Springer 2013, Berlin, Heidelberg, pp 174–181. https://doi.org/10.1007/978-3-642-38899-6_21
Zhuang X, Hawkes DJ, Crum WR, Boubertakh R, Uribe S, Atkinson D (2008) Robust registration between cardiac MRI images and atlas for segmentation propagation. International Society for Optics and Photonics 6914:547–550. https://doi.org/10.1117/12.769445
Kirisli HA, Schaap M, Klein S, Neefjes LA, Weustink AC, Van Walsum T (2010) Fully automatic cardiac segmentation from 3D CTA data: a multi-atlas based approach. Med Imaging 7623. International Society for Optics and Photonics 2010:762305. https://doi.org/10.1117/12.838370
Bai W, Shi W, O’Regan DP et al (2013) A probabilistic patch-based label fusion model for multi-atlas segmentation with registration refinement: application to cardiac MR images. IEEE Trans Med Imaging 32:1302–1315. https://doi.org/10.1016/J.MEDIA.2004.06.005
Zhuang X, Rhode KS, Razavi RS et al (2010) A registration-based propagation framework for automatic whole heart segmentation of cardiac MRI. IEEE T Med Imaging 29:1612–1625. https://doi.org/10.1109/TMI.2010.2047112
Grosgeorge D, Petitjean C, Dacher J-N, Ruan S (2013) Graph cut segmentation with a statistical shape model in cardiac MRI. Comput Vis Image Underst 117:1027–1035. https://doi.org/10.1016/J.CVIU.2013.01.014
Peng B, Zhang L, Zhang D (2013) A survey of graph theoretical approaches to image segmentation. Pattern Recogn 46:1020–1038. https://doi.org/10.1016/J.PATCOG.2012.09.015
Lu XS, Ma S, Zhao DQ, LM (2017) Graph cuts segmentation based on multi-dimensional features for the right ventricle in cardiac MRI. J Eng 2017:123–125. https://doi.org/10.1049/joe.2016.0288
Russakovsky O, Deng J, Su H, Krause J, Satheesh S, Ma S, Huang Z, Karpathy A, Khosla A, Bernstein M (2015) Imagenet large scale visual recognition challenge. Int J Comput Vis 115:211–252. https://doi.org/10.1007/s11263-015-0816-y
Krizhevsky A, Sutskever I, Hinton G (2012) Imagenet classification with deep convolutional neural networks. Neural Information Processing Systems 25. https://doi.org/10.1145/3065386
Avendi MR, Kheradvar A, Jafarkhani H (2017) Automatic segmentation of the right ventricle from cardiac MRI using a learning-based approach. Magn Reson Med 78:2439–2448. https://doi.org/10.1002/mrm.26631
Zotti C, Luo Z, Lalande A et al (2018) Convolutional neural network with shape prior applied to cardiac MRI segmentation. IEEE J Biomed Health 2018:1–1. https://doi.org/10.1109/JBHI.2018.2865450
Vigneault DM, Xie WD, Ho CY, Bluemke DA, Noble JA (2018) Ω-Net (Omega-Net): fully automatic, multi-view cardiac MR detection, orientation, and segmentation with deep neural networks. Med Image Anal 2018:95–106. https://doi.org/10.1016/j.media.2018.05.008
Long J, Shelhamer E, Darrell T (2015) Fully convolutional networks for semantic segmentation. 2015 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Boston, MA 2015:3431–3440. https://doi.org/10.1109/CVPR.2015.7298965
Tran PV (2016) A fully convolutional neural network for cardiac segmentation in short-axis MRI. arXiv preprint arXiv 1604:00494
Khened M, Alex V, Krishnamurthi G (2018) Fully convolutional multi-scale residual DenseNets for cardiac segmentation and automated cardiac diagnosis using ensemble of classifiers. Med Image Anal 51:21–45. https://doi.org/10.1016/j.media.2018.10.004
Avendi MR, Kheradvar A, Jafarkhani H (2016) A combined deep-learning and deformable-model approach to fully automatic segmentation of the left ventricle in cardiac MRI. Med Image Anal 30:108–119. https://doi.org/10.1016/j.media.2016.01.005
Patravali J, Jain S, Chilamkurthy S (2017) 2D-3D Fully convolutional neural networks for cardiac MR segmentation. International Workshop on Statistical Atlases and Computational Models of the Heart 2017:130–139
Jégou S, Drozdzal M, Vazquez D et al (2017) The one hundred layers tiramisu: fully convolutional DenseNets for semantic segmentation. IEEE Conference on Computer Vision and Pattern Recognition Workshops 2017:1175–1183. https://doi.org/10.1109/CVPRW.2017.156
Li C, Huang R, Ding Z et al (2011) A level set method for image segmentation in the presence of intensity inhomogeneities with application to MRI. IEEE T Image Process 20:2007–2016. https://doi.org/10.1109/TIP.2011.2146190
Huang G, Liu Z, Weinberger KQ, van der Maaten L (2017) Densely connected convolutional networks. Proc IEEE Conf Comput Vis Pattern Recognit 2017:4700-4708. https://doi.org/10.1109/CVPR.2017.243
Ioffe S, Szegedy C (2015) Batch normalization: accelerating deep network training by reducing internal covariate shift. arXiv preprint arXiv 1502:03167
Glorot X, Bordes A, Bengio Y (2011) Deep sparse rectifier neural networks. Proceedings of the 14th International Conference on Artificial Intelligence and Statistics (AISTATS) 315:323
Szegedy C, Vanhoucke V, Ioffe S, Shlens J, Wojna Z (2016) Rethinking the inception architecture for computer vision. In Conference on Computer Vision & Pattern Recognition IEEE Computer Society. https://doi.org/10.1109/CVPR.2016.308
He K, Zhang X, Ren S, Sun J (2016) Deep residual learning for image recognition. In: Conference on Computer Vision & Pattern Recognition IEEE Computer Society. https://doi.org/10.1109/CVPR.2016.90
Petitjean C, Zuluaga MA, Bai W, Dacher J-N, Grosgeorge D, Caudron J et al (2015) Right ventricle segmentation from cardiac MRI: a collation study. Med Image Anal 19:187–202. https://doi.org/10.1016/J.MEDIA.2014.10.004
Ronneberger O, Fischer P, Brox T (2015) U-Net: Convolutional networks for biomedical image segmentation. International Conference on Medical Image Computing and Computer-Assisted Intervention 2015:234–241. https://doi.org/10.1007/978-3-319-24574-4_28
Luo Y, Yang B, Xu LS et al (2018) Segmentation of the left ventricle in cardiac MRI using a hierarchical extreme learning machine model. Int J Mach Learn Cybern 9:1741–1751. https://doi.org/10.1007/s13042-017-0678-4
Acknowledgments
Many thanks to Prof. Stephen E. Greenwald (from School of Medicine and Dentistry, Queen Mary, University of London, UK) for reviewing the manuscript, improving the English, and providing helpful suggestions. We gratefully acknowledge the kind assistance of Prof. Benqiang Yang and Dr. Junrui Xiao in data collection and delineating the entire image dataset.
Funding
The research reported here was, in part, supported by the National Natural Science Foundation of China (No. 61773110, No. 61374015), the Natural Science Foundation of Liaoning Province (No. 20170540312), and the Fundamental Research Funds for the Central Universities (Nos. N181906001 and N181604006). This research is also supported by the Shenyang Science and Technology Plan Fund (No. 20-201-4-10), the Member Program of Neusoft Research of Intelligent Healthcare Technology, Co. Ltd. (No. MCMP062002).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Luo, Y., Xu, L. & Qi, L. A cascaded FC-DenseNet and level set method (FCDL) for fully automatic segmentation of the right ventricle in cardiac MRI. Med Biol Eng Comput 59, 561–574 (2021). https://doi.org/10.1007/s11517-020-02305-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11517-020-02305-7