Abstract
For cancer prediction, the prognostic stage is the main factor that helps medical experts to decide the optimal treatment for a patient. Specialists study prognostic stage information from medical reports, often in an unstructured form, and take a larger review time. The main objective of this study is to suggest a generic clinical decision-unifying staging method to extract the most reliable prognostic stage information of breast cancer from medical records of various health institutions. Additional prognostic elements should be extracted from medical reports to identify the cancer stage for getting an exact measure of cancer and improving care quality. This study has collected 465 pathological and clinical reports of breast cancer sufferers from India’s reputed medical institutions. The unstructured records were found distinct from each institute. Anatomic and biologic factors are extracted from medical records using the natural language processing, machine learning and rule-based method for prognostic stage detection. This study has extracted anatomic stage, grade, estrogen receptor (ER), progesterone receptor (PR), and human epidermal growth factor receptor 2 (HER2) from medical reports with high accuracy and predicted prognostic stage for both regions. The prognostic stage prediction’s average accuracy is found 92% and 82% in rural and urban areas, respectively. It was essential to combine biological and anatomical elements under a single prognostic staging method. A generic clinical decision-unifying staging method for prognostic stage detection with great accuracy in various institutions of different regional areas suggests that the proposed research improves the prognosis of breast cancer.
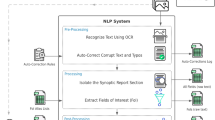
Graphical abstract
















Similar content being viewed by others
References
Cancer Statistics in India. http://cancerindia.org.in/cancer-statistics/. Accessed 25 Nov 2020
Mathur P, Sathishkumar K, Chaturvedi M, Das P, Sudarshan K, Santhappan S, Nallasamy V, John A, Narasimhan S, Roselind F (2020) Cancer Statistics, 2020: Report from National Cancer Registry Programme, India. JCO Global Oncol 6:1063–1075. https://doi.org/10.1200/GO.20.00122
Martinez D, Cavedon L, Pitson G (2013) Stability of text mining techniques for identifying cancer staging. In: Louhi, The 4th International Workshop on Health Document Text Mining and Information Analysis, NICTA, Canberra, Australia
Kim BJ, Merchant M, Zheng C, Thomas AA, Contreras R, Jacobsen SJ, Chien GW (2014) Second prize: “A natural language processing program effectively extracts key pathologic findings from radical prostatectomy reports.” J Endourol 28(12):1474–1478. https://doi.org/10.1089/end.2014.0221
Wen-wai Y, Meliha Y (2016) Natural Language Processing in Oncology a Review. J Am Med Inform Assoc 2(6):797–804. https://doi.org/10.1001/jamaoncol.2016.0213
Cheng LTE, Zheng J, Savova GK, Erickson BJ (2010) Discerning tumor status from unstructured MRI reports: completeness of information in existing reports and utility of automated natural language processing. J Digit Imaging 23(2):119–132. https://doi.org/10.1007/s10278-009-9215-7
Edge SB, Byrd DR, Compton CC, Fritz AG, Greene FL, Trotti A (2011) AJCC cancer staging manual, 7th edn. Springer-Verlag, Berlin. ISBN 978-0-387-88440-0
Spasic I, Livsey J, Keane JA, Nenadic G (2014) Text mining of cancer-related information: Review of current status and future directions. Int J Med Informatics 83:605–623. https://doi.org/10.1016/j.ijmedinf.2014.06.009
Deshmukh PR, Phalnikar R (2020) TNM cancer stage detection from unstructured pathology reports of breast cancer patients. In: Bhalla S et al (eds) Proceeding of International conference on computational science and applications, algorithms for intelligent systems. Springer Nature Singapore Pte Ltd., CH 40:411–418. https://doi.org/10.1007/978-981-15-0790-8_40
Ravi K, Ramachandra GA, Nagamani K (2013) An Efficient Prediction of Breast Cancer Data using Data Mining Techniques. Int J Innov Eng Technol 2(4):139–144. SSN: 2319-1058
Chatterjee S, Chattopadhayay A (2016) Cancer Registration in India– Current Scenario and Future Perspectives. Asian Pac J Cancer Prev 17(8):3687–3696. https://doi.org/10.14456/apjcp.2016.154/APJCP.2016.17.8.3687
Wong RX, Wong FY, Lim J, Lian WX, Yap YS (2018) Validation of the AJCC 8th prognostic system for breast cancer in an Asian healthcare setting. Breast 40:38–44. https://doi.org/10.1016/j.breast.2018.04.013. Elsevier
Wang M, Chen H, Kejin W, Ang D, Mingdi Z, Peng Z (2018) Evaluation of the prognostic stage in the 8th edition of the American Joint Committee on Cancer in locally advanced breast cancer: An analysis based on SEER 18 database. Breast 37:56–63. https://doi.org/10.1016/j.breast.2017.10.011
National centre for Disease Informatics and Research, National Cancer Registry Program, http://www.ncrpindia.org/. Accessed 25 Nov 2020
Yokoyama S, Hamada T, Higashi M, Matsuo K, Maemura K, Kurahara H, Horinouchi M, Hiraki T, Sugimoto T, Akahane T, Yonezawa S, Kornmann M, Batra SK, Hollingsworth MA, Tanimoto A (2020) Predicted Prognosis of Patients with Pancreatic Cancer by Machine Learning. Clin Cancer Res 26:2411–2421. https://doi.org/10.1158/1078-0432,January28
Li J, Li Z, Luo J, Yao Y (2020) ACNNT3: Attention-CNN Framework for Prediction of Sequence- Based Bacterial Type III Secreted Effectors. Comput Math Methods Med Article ID 3974598:7. https://doi.org/10.1155/2020/3974598
Li Z, Zhu J, Xu X, Yao Y (2020) RDense: a protein-RNA binding prediction model based on bidirectional recurrent neural network and densely connected convolutional networks. IEEE Access 8. https://doi.org/10.1109/ACCESS.2019.2961260.
Jiang X, Zhao J, Qian W, Song W, Ning LG (2020) A generative adversarial network model for disease gene prediction with RNA-seq data. IEEE Access 8. https://doi.org/10.1109/ACCESS.2020.2975585.
Mignone P, Pio G, D’Elia D, Ceci M (2020) Exploiting transfer learning for the reconstruction of the human gene regulatory network. Bioinformatics 36(5):1553–1561. https://doi.org/10.1093/bioinformatics/btz781
Pio G, Ceci M, Prisciandaro F, Malerba D (2020) Exploiting causality in gene network reconstruction based on graph embedding. Mach Learn 109:1231–1279. https://doi.org/10.1007/s10994-019-05861-8
Barracchia EP, Pio G, Delia D, Ceci M (2020) Prediction of new associations between ncRNAs and diseases exploiting multi-type hierarchical clustering. BMC Bioinformatics 21:70. https://doi.org/10.1186/s12859-020-3392-2
Jiang D, Liao J, Duan H, Wu Q, Owen G, Shu C, Chen L, He Y, Wu Z, He D, Zhang W, Wang Z (2020) A machine learning-based prognostic predictor for stage III colon cancer. Sci Rep 10:10333. https://doi.org/10.1038/s41598-020-67178-0
Muhammad A, Maqbool H, Wajahat Ali K, Ali T, Lee S, Huh E-N, Hafiz Farooq A, Arif J, Hassan I, Muhammad I, Manzar Abbas H (2017) Comprehensible knowledge model creation for cancer treatment decision making. Comput Biol Med 82:119–129. https://doi.org/10.1016/j.compbiomed.2017.01.010. Science Direct, Elsevier
Martinez D, Pitson G, MacKinlay A, Cavedon L (2014) Cross-hospital portability of information extraction of cancer staging information. Artif Intell Med 62:11–21. https://doi.org/10.1016/j.artmed.2014.06.002. Elsevier
Nguyen AN, Lawley MJ, Hansen DP, Bowman RV, Clarke BE, Duhig EE, Colquist S (2010) Symbolic rule-based classification of lung cancer stages from free-text pathology reports. J Am Med Inform Assoc 17:440–445. https://doi.org/10.1136/jamia.2010.003707
Rani GJJ, Gladis D, Mammen JJ (2017) Comparison of breast cancer staging in natural language text and SNOMED annotated text. Int J Pure Appl Math 116(21):243–249
Warner JL, Mia AL, Michael NN (2016) Feasibility and accuracy of extracting cancer stage information from narrative electronic health record data. Am Soc Clin Oncol 12(2). https://doi.org/10.1200/JOP.2015.004622.
Martinez D, Li Y (2011) Information extraction from pathology reports in a Hospital setting. CIKM’11, 1877–1882, ACM 978-1-4503-0717-8/11/10, October 24–28
McCowan I, Moore D, Fry M-J (2006) Classification of cancer stage from free-text histology reports. International Conference of the IEEE Engineering in Medicine and Biology Society. https://doi.org/10.1109/IEMBS.2006.259563
Rani GJJ, Gladis D, Mammen JJ (2019) SNOMED CT annotation for improved pathological decisions in breast cancer domain. Int J Recent Technol Eng 8(3). https://doi.org/10.35940/ijrte.C6519.098319
Nguyen A, Moore D, McCowan I, Courage M Multi-class classification of cancer stages from free-text histology reports using support vector machines. 29th Annual International Conference of the IEEE EMBS, France IEEE 2007, pp 5140–5143, https://doi.org/10.1109/IEMBS.2007.4353497
Rajaguru H, Vasanthi NS, Balasubramani M (2012) Performance analysis of artificial neural networks and statistical methods in classification of oral and breast cancer stages. Int J Soft Comput Eng 2(3)
McCowan IA, Moore DC, Nguyen AN, Bowman RV, Clarke BE, Duhig EE, Fry M-J (2007) Collection of cancer stage data by classifying free-text medical reports. J Am Med Inform Assoc 14(6):736–745. https://doi.org/10.1197/jamia.M2130
Dursun D, Glenn W, Amit K (2005) Predicting breast cancer survivability: a comparison of three data mining methods. Artif Intell Med 34:113–127. https://doi.org/10.1016/j.artmed.2004.07.002. Elsevier
Joseph AC, David SW (2006) Applications of machine learning in cancer prediction and prognosis. Cancer Informat 2:59–77. PMID: 19458758, PMCID: PMC2675494
Dechang C, Huan W, Li S, Matthew TH, Donald EH, Arnold MS, Jigar AP (2016) An algorithm for creating prognostic systems for cancer. J Med Syst 40:160. https://doi.org/10.1007/s10916-016-0518-1. Springer
Deshmukh PR, Phalnikar R Identifying contextual information in medical document classification using term weighting. IEEE 8th International Advanced Computing Conference at Bennett University, Greater Noida, India, 17th -18th Dec 2018
U.S. National Library of Medicine (2008) Unified medical language system (UMLS). https://www.nlm.nih.gov/research/umls/. Accessed 25 Nov 2020
Chapman WW, Bridewell W, Hanbury P, Cooper GF, Buchanan BG (2001) A simple algorithm for identifying negated findings and diseases in discharge sum-maries. J Biomed Inform 34(5):301–310. https://doi.org/10.1006/jbin.2001.1029
www.Breastcancer.org. Accessed 25 Nov 2020
Sanjay PB, Partha SR, Myung-Shin S, Xing Y, Jaime MS, Xiaojiang C, Armando EG (2014) Personalizing breast cancer staging by the inclusion of ER, PR, and HER2. JAMA 149(2):125–129. https://doi.org/10.1001/jamasurg.2013.3181
Buckley JM, Coopey SB, Sharko J (2012) The feasibility of using natural language processing to extract clinical information from breast pathology reports. J Pathol Inform 3:23. https://doi.org/10.4103/2153-3539.97788
Dixit A, Singh R (2017) Multiple sliding window based pattern matching algorithms: survey. International Journal of Creative Research Thoughts (IJCRT) 5(4):3453–3458
Amjad H, Rola A, Dima S (2015) Four sliding windows pattern matching algorithms. J Softw Eng Appl. https://doi.org/10.4236/jsea.2015.83016
Hortobagyi GN, Connolly JL, D’Orsi CJ, Edge SB, Mittendorf EA, Rugo HS, Solin LJ, Weaver DL, Winchester DJ, Giuliano A AJCC Cancer staging manual eighth edition. https://doi.org/10.1007/978-3-319-40618-3_48
Mogana DG, Nur AT, Yip CH, Pietro L, Sarinder KD (2019) Predicting factors for survival of breast cancer patients using machine learning Techniques. BMC Med Inform Decis Mak 19:48. https://doi.org/10.1186/s12911-019-0801-4
Acknowledgements
The authors would like to convey thanks to Nurgis Dutta Memorial Cancer Hospital (NDMCH) in the rural region and Jehangir Hospital (JH) and different laboratories in the urban area for allowing access to their medical records. The authors also want to acknowledge Ethics Committees for approving proposed research. The authors would like to express their gratitude to Dr. Nene, Dr. Chauhan, Dr. Joshi, Dr. Shilpa, Dr. Vibhute, and Dr. Mane for their guidance in the medical field. Thanks to all patients with breast cancer for letting the authors use their medical records for this research. The authors would like to thank Harshal Ingale for his cooperation.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Deshmukh, P.R., Phalnikar, R. Information extraction for prognostic stage prediction from breast cancer medical records using NLP and ML. Med Biol Eng Comput 59, 1751–1772 (2021). https://doi.org/10.1007/s11517-021-02399-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11517-021-02399-7