Abstract
Purpose
Radiological longitudinal follow-up of liver tumors in CT scans is the standard of care for disease progression assessment and for liver tumor therapy. Finding new tumors in the follow-up scan is essential to determine malignancy, to evaluate the total tumor burden, and to determine treatment efficacy. Since new tumors are typically small, they may be missed by examining radiologists.
Methods
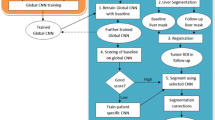
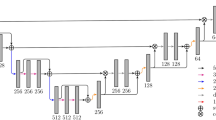
We describe a new method for the automatic detection and segmentation of new tumors in longitudinal liver CT studies and for liver tumors burden quantification. Its inputs are the baseline and follow-up CT scans, the baseline tumors delineation, and a tumor appearance prior model. Its outputs are the new tumors segmentations in the follow-up scan, the tumor burden quantification in both scans, and the tumor burden change. Our method is the first comprehensive method that is explicitly designed to find new liver tumors. It integrates information from the scans, the baseline known tumors delineations, and a tumor appearance prior model in the form of a global convolutional neural network classifier. Unlike other deep learning-based methods, it does not require large tagged training sets.
Results
Our experimental results on 246 tumors, of which 97 were new tumors, from 37 longitudinal liver CT studies with radiologist approved ground-truth segmentations, yields a true positive new tumors detection rate of 86 versus 72% with stand-alone detection, and a tumor burden volume overlap error of 16%.
Conclusions
New tumors detection and tumor burden volumetry are important for diagnosis and treatment. Our new method enables a simplified radiologist-friendly workflow that is potentially more accurate and reliable than the existing one by automatically and accurately following known tumors and detecting new tumors in the follow-up scan.






Similar content being viewed by others
References
Eisenhauer E, Therasse P, Bogaerts J, Schwartz LH, Sargent D, Ford R, Dancey J, Arbuck S, Gwyther S, Mooney M, Rubinstein L (2009) New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur J Cancer 45(2):228–247
Tuma RS (2006) Sometimes size doesn’t matter: reevaluating RECIST and tumor response rate endpoints. J Nat Cancer Inst 98:1272–1274
Vivanti R, Ephrat A, Joskowicz L, Lev-Cohain N, Karaaslan OA, Sosna J (2015) Automatic liver tumor segmentation in follow-up CT scans: preliminary method and results. In: International workshop on patch-based techniques in medical imaging, pp 54–61
Deng X, Du G (2008) 3D segmentation in the clinic: a grand challenge II-liver tumor segmentation. In: MICCAI workshop
Moltz J, Bornemann L, Dicken V, Peitgen H (2008) Segmentation of liver metastases in CT scans by adaptive thresholding and morphological processing. In: Proceedings of the MICCAI workshop on 3D segmentation in the clinic: a Grand Challenge II—Liver tumor segmentation
Shimizu A, Narihira T, Furukawa D, Kobatake H, Nawano S, Shinozaki K (2008) Ensemble segmentation using AdaBoost with application to liver lesion extraction from a CT volume. In: Proceedings of the MICCAI workshop on 3D segmentation in the clinic: a Grand challenge II—Liver tumor segmentation
Luo S, Li X, Li J (2014) Review on the methods of automatic liver segmentation from abdominal images. J Comput Commun 2(2):1
Hong JS, Kaneto T, Sekiguchi R, Park KH (2001) Automatic liver tumor detection from CT. IEEE Trans Inf Syst 84(6):741–748
Chen EL (1998) An automatic diagnostic system for CT liver image classification. IEEE Trans Biomed Eng 45(6):783–794
Li W, Jia F, Hu Q (2015) Automatic segmentation of liver tumors in CT images with deep convolutional neural networks. J Comput Commun 3(11):146
Ben-Cohen A, Diamant I, Klang E, Amitai M, Greenspan H (2016) Fully convolutional network for liver segmentation and lesions detection. In: Proceedings of the international workshop on large-scale annotation of biomedical data and expert label synthesis. Springer, pp 77–85
Christ PF, Elshaer ME, Ettlinger F, Tatavarty S, Bickel M, Bilic P, Rempfler M, Armbruster M, Hofmann F, D’Anastasi M, Sommer WH (2016) Automatic liver and lesion segmentation in CT using cascaded fully convolutional neural networks and 3D conditional random fields. In: Proceedings of the international conference on medical image computing and computer-assisted intervention. Springer International Publishing, pp 415–423
Bilello M, Gokturk SB, Desser T, Napel S, Jeffrey RB Jr, Beaulieu CF (2004) Automatic detection and classification of hypodense hepatic lesions on contrast-enhanced venous-phase CT. Med Phys 31(9):2584–93
Masuda Y, Foruzan AH, Tateyama T, Chen YW (2010) Automatic liver tumor detection using EM/MPM algorithm and shape information. In: Proceedings of the 2nd IEEE international conference on software engineering and data mining (SEDM), pp 692–695
Lu D, Mausel P, Brondizio E, Moran E (2004) Change detection techniques. Int J Remote Sens 25(12):2365–401
Weizman L, Ben-Sira L, Joskowicz L, Precel R, Constantini S, Ben-Bashat D (2010) Automatic segmentation and components classification of optic pathway gliomas in MRI. Med Image Comput Comput-Assist Interv 1:103–110
Vivanti R, Joskowicz L, Karaaslan OA, Sosna J (2015) Automatic lung tumor segmentation with leaks removal in follow-up CT studies. Int J Comput Assist Radiol Surg 10(9):1505–1514
Ben-Cohen A, Diamant I, Klang E, Amitai M, Greenspan H (2014) Automatic detection and segmentation of liver metastatic lesions on serial CT examinations. In: Proceedings of the SPIE medical imaging conference, pp 903519–903527
Glocker B, Sotiras A, Komodakis N, Paragios N (2011) Deformable medical image registration: setting the state of the art with discrete methods. Ann Rev Biomed Eng 13:219–44
Chan TF, Vese LA (2001) Active contours without edges. IEEE Trans Image Process 10(2):266–77
Liaw A, Wiener M (2002) Classification and regression by random forest. Radiol News 2(3):18–22
Joachims, T (1998) Making large-scale SVM learning practical. Technical Report SFB 475: Komplexitätsreduktion in Multivariaten Datenstrukturen, Universität Dortmund
Chen T, Guestrin, C (2016) Xgboost: a scalable tree boosting system. In: Proceedings of the 22nd ACM SIGKDD international conference on knowledge discovery and data mining, pp 345–364
www.mathworks.com/matlabcentral/fileexchange/23847-sparse-field-methods-for-active-contours
Acknowledgements
This work was partially supported by Grant 53681 from the Israel Ministry of Science, Technology and Space entitled: METASEG: a new medical image segmentation paradigm for clinical decision support and big data radiology.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
None of the authors has any conflict of interest. The authors have no personal financial or institutional interest in any of the materials, software or devices described in this article.
Protection of human and animal rights statement
No animals or humans were involved in this research. All scans were anonymized before delivery to the researchers.
Rights and permissions
About this article
Cite this article
Vivanti, R., Szeskin, A., Lev-Cohain, N. et al. Automatic detection of new tumors and tumor burden evaluation in longitudinal liver CT scan studies. Int J CARS 12, 1945–1957 (2017). https://doi.org/10.1007/s11548-017-1660-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11548-017-1660-z