Abstract
Purpose
Most of the existing convolutional neural network (CNN)-based medical image segmentation methods are based on methods that have originally been developed for segmentation of natural images. Therefore, they largely ignore the differences between the two domains, such as the smaller degree of variability in the shape and appearance of the target volume and the smaller amounts of training data in medical applications. We propose a CNN-based method for prostate segmentation in MRI that employs statistical shape models to address these issues.
Methods
Our CNN predicts the location of the prostate center and the parameters of the shape model, which determine the position of prostate surface keypoints. To train such a large model for segmentation of 3D images using small data (1) we adopt a stage-wise training strategy by first training the network to predict the prostate center and subsequently adding modules for predicting the parameters of the shape model and prostate rotation, (2) we propose a data augmentation method whereby the training images and their prostate surface keypoints are deformed according to the displacements computed based on the shape model, and (3) we employ various regularization techniques.
Results
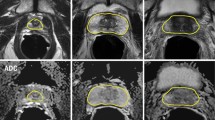
Our proposed method achieves a Dice score of 0.88, which is obtained by using both elastic-net and spectral dropout for regularization. Compared with a standard CNN-based method, our method shows significantly better segmentation performance on the prostate base and apex. Our experiments also show that data augmentation using the shape model significantly improves the segmentation results.
Conclusions
Prior knowledge about the shape of the target organ can improve the performance of CNN-based segmentation methods, especially where image features are not sufficient for a precise segmentation. Statistical shape models can also be employed to synthesize additional training data that can ease the training of large CNNs.



Similar content being viewed by others
References
Chen L, Papandreou G, Kokkinos I, Murphy K, Yuille AL (2016) Deeplab: semantic image segmentation with deep convolutional nets, atrous convolution, and fully connected crfs. CoRR abs/1606.00915. http://arxiv.org/abs/1606.00915
Havaei M, Davy A, Warde-Farley D, Biard A, Courville A, Bengio Y, Pal C, Jodoin PM, Larochelle H (2017) Brain tumor segmentation with deep neural networks. Med Image Anal 35:18–31
He K, Zhang X, Ren S, Sun J (2015) Delving deep into rectifiers: surpassing human-level performance on imagenet classification. In: IEEE international conference on computer vision (ICCV) 2015
He K, Zhang X, Ren S, Sun J (2016) Deep residual learning for image recognition. In: IEEE conference on computer vision and pattern recognition (CVPR), 2016
Heimann T, Meinzer HP (2009) Statistical shape models for 3d medical image segmentation: a review. Med Image Anal 13(4):543–563. https://doi.org/10.1016/j.media.2009.05.004
Khan S, Hayat M, Porikli F (2017) Regularization of deep neural networks with spectral dropout. arXiv preprint arXiv:1711.08591
Klein S, van der Heide UA, Lips IM, van Vulpen M, Staring M, Pluim JPW (2008) Automatic segmentation of the prostate in 3d mr images by atlas matching using localized mutual information. Med. Phys. 35(4):1407–1417. https://doi.org/10.1118/1.2842076
Li W, Liao S, Feng Q, Chen W, Shen D (2011) Learning image context for segmentation of prostate in CT-guided radiotherapy. Springer, Berlin, Heidelberg, pp 570–578
Litjens G, Kooi T, Bejnordi BE, Setio AAA, Ciompi F, Ghafoorian M, van der Laak JA, van Ginneken B, Snchez CI (2017) A survey on deep learning in medical image analysis. Med Image Anal 42(Supplement C):60–88. https://doi.org/10.1016/j.media.2017.07.005
Litjens G, Toth R, van de Ven W, Hoeks C, Kerkstra S, van Ginneken B, Vincent G, Guillard G, Birbeck N, Zhang J, Strand R, Malmberg F, Ou Y, Davatzikos C, Kirschner M, Jung F, Yuan J, Qiu W, Gao Q, Edwards PE, Maan B, van der Heijden F, Ghose S, Mitra J, Dowling J, Barratt D, Huisman H, Madabhushi A (2014) Evaluation of prostate segmentation algorithms for mri: the promise12 challenge. Med Image Anal 18(2):359–373. https://doi.org/10.1016/j.media.2013.12.002
Long J, Shelhamer E, Darrell T (2015) Fully convolutional networks for semantic segmentation. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 3431–3440
Mahapatra D, Buhmann JM (2014) Prostate mri segmentation using learned semantic knowledge and graph cuts. IEEE Trans Biomed Eng 61(3):756–764. https://doi.org/10.1109/TBME.2013.2289306
Makni N, Puech P, Lopes R, Dewalle AS, Colot O, Betrouni N (2008) Combining a deformable model and a probabilistic framework for an automatic 3d segmentation of prostate on mri. Int J Comput Assisted Radiol Surg 4(2):181. https://doi.org/10.1007/s11548-008-0281-y
Martin S, Troccaz J, Daanen V (2010) Automated segmentation of the prostate in 3d mr images using a probabilistic atlas and a spatially constrained deformable model. Med Phys 37(4):1579–1590. https://doi.org/10.1118/1.3315367
Milletari F, Navab N, Ahmadi SA (2016) V-net: fully convolutional neural networks for volumetric medical image segmentation. In: Fourth international conference on 3D vision (3DV), 2016, pp 565–571. IEEE
Milletari F, Rothberg A, Jia J, Sofka M (2017) Integrating statistical prior knowledge into convolutional neural networks. Springer, Cham, pp 161–168
Myronenko A, Song X (2010) Point set registration: coherent point drift. IEEE Trans Pattern Anal Mach Intell 32(12):2262–2275
Ravishankar H, Venkataramani R, Thiruvenkadam S, Sudhakar P, Vaidya V (2017) Learning and incorporating shape models for semantic segmentation. Springer, Cham, pp 203–211
Ren S, He K, Girshick R, Sun J (2015) Faster r-cnn: towards real-time object detection with region proposal networks. In: Advances in neural information processing systems, pp 91–99
Ronneberger O, Fischer P, Brox T (2015) U-net: Convolutional networks for biomedical image segmentation. In: International conference on medical image computing and computer-assisted intervention, pp 234–241. Springer
Sciolla B, Martin M, Delachartre P (2017) Multi-pass 3d convolutional neural network segmentation of prostate mri images
Srivastava N, Hinton GE, Krizhevsky A, Sutskever I, Salakhutdinov R (2014) Dropout: a simple way to prevent neural networks from overfitting. J Mach Learn Res 15(1):1929–1958
Toth R, Madabhushi A (2012) Multifeature landmark-free active appearance models: application to prostate mri segmentation. IEEE Trans Med Imaging 31(8):1638–1650
Yu L, Yang X, Chen H, Qin J, Heng PA (2017) Volumetric convnets with mixed residual connections for automated prostate segmentation from 3d mr images
Zou H, Hastie T (2005) Regularization and variable selection via the elastic net. J R Stat Soc Ser B (Stat Methodol) 67(2):301–320
Acknowledgements
This project is funded by the Canadian Institutes of Health Research (CIHR) and the Prostate Cancer Canada (PCC). We would like to thank the support from the Charles Laszlo Chair in Biomedical Engineering held by Professor Salcudean. The authors also gratefully acknowledge the help from physicians and staff at the Vancouver Cancer Centre who have contributed to this project (Grant Nos. MOP-142439, D2016-1352).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Rights and permissions
About this article
Cite this article
Karimi, D., Samei, G., Kesch, C. et al. Prostate segmentation in MRI using a convolutional neural network architecture and training strategy based on statistical shape models. Int J CARS 13, 1211–1219 (2018). https://doi.org/10.1007/s11548-018-1785-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11548-018-1785-8