Abstract
Purpose
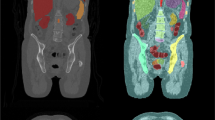
Automated segmentation of torso organs from positron emission tomography/computed tomography (PET/CT) images is a prerequisite step for nuclear medicine image analysis. However, accurate organ segmentation from clinical PET/CT is challenging due to the poor soft tissue contrast in the low-dose CT image and the low spatial resolution of the PET image. To overcome these challenges, we developed a multi-atlas segmentation (MAS) framework for torso organ segmentation from 2-deoxy-2-[18F]fluoro-d-glucose PET/CT images.
Method
Our key idea is to use PET information to compensate for the imperfect CT contrast and use surface-based atlas fusion to overcome the low PET resolution. First, all the organs are segmented from CT using a conventional MAS method, and then the abdomen region of the PET image is automatically cropped. Focusing on the cropped PET image, a refined MAS segmentation of the abdominal organs is performed, using a surface-based atlas fusion approach to reach subvoxel accuracy.
Results
This method was validated based on 69 PET/CT images. The Dice coefficients of the target organs were between 0.80 and 0.96, and the average surface distances were between 1.58 and 2.44 mm. Compared to the CT-based segmentation, the PET-based segmentation gained a Dice increase of 0.06 and an ASD decrease of 0.38 mm. The surface-based atlas fusion leads to significant accuracy improvement for the liver and kidneys and saved ~ 10 min computation time compared to volumetric atlas fusion.
Conclusions
The presented method achieves better segmentation accuracy than conventional MAS method within acceptable computation time for clinical applications.




Similar content being viewed by others
References
Jimenez-Del-Toro O, Muller H, Krenn M, Gruenberg K, Taha AA, Winterstein M, Eggel I, Foncubierta-Rodriguez A, Goksel O, Jakab A (2016) Cloud-based evaluation of anatomical structure segmentation and landmark detection algorithms: VISCERAL anatomy benchmarks. IEEE Trans Med Imag 35(11):2459–2475
Norajitra T, Maier-Hein KH (2017) 3D statistical shape models incorporating landmark-wise random regression forests for omni-directional landmark detection. IEEE Trans Med Imaging 36(1):155–168
Criminisi A, Robertson D, Konukoglu E, Shotton J, Pathak S, White S, Siddiqui K (2013) Regression forests for efficient anatomy detection and localization in computed tomography scans. Med Image Anal 17(8):1293–1303
Zhou X, Wang S, Chen H, Hara T, Yokoyama R, Kanematsu M, Fujita H (2012) Automatic localization of solid organs on 3D CT images by a collaborative majority voting decision based on ensemble learning. Comput Med Imag Gr Off J Comput Med Imag Soc 36(4):304–313
Seifert S (2010) Semantic annotation of medical images. Acta Biol Colomb 15(3):181–196
Seifert S, Barbu A, Feulner J, Suehling M (2008) Hierarchical parsing and semantic navigation of full body CT data. Proc SPIE 2008:725902–725908
Zhou X, Watanabe A, Zhou X, Hara T, Yokoyama R, Kanematsu M, Fujita H (2012) Automatic organ segmentation on torso CT images by using content-based image retrieval. Proc SPIE Int Soc Opt Eng 8314:116–123
Udupa JK, Odhner D, Zhao L, Tong Y, Matsumoto MMS, Ciesielski KC, Falcao AX, Vaideeswaran P, Ciesielski V, Saboury B (2014) Body-Wide hierarchical fuzzy modeling, recognition, and delineation of anatomy in medical images. Med Image Anal 18(5):752–771
Wang CSO (2014) Automatic multi-organ segmentation using fast model based level set method and hierarchical shape priors. Proc VISC Chall ISBI 1194:25–31
Wang H, Udupa JK, Odhner D, Tong Y, Zhao L, Torigian DA (2016) Automatic anatomy recognition in whole-body PET/CT images. Med Phys 43(1):613–629
Lay N, Birkbeck N, Zhang J, Zhou SK (2013) Rapid multi-organ segmentation using context integration and discriminative models. Int Conf Inf Process Med Imag (IPIM), 2013. Springer, Berlin, pp 450–462
Gauriau R, Ardori R, Lesage D, Bloch I (2015) Multiple template deformation application to abdominal organ segmentation. In: IEEE international symposium on biomedical imaging, pp 359–362
Bagci U, Chen X, Udupa JK (2012) Hierarchical scale-based multiobject recognition of 3-D anatomical structures. IEEE Trans Med Imag 31(3):777–789
Chen X, Udupa JK, Bağcı U, Ying Z, Yao J (2012) Medical image segmentation by combining graph cut and oriented active appearance models. IEEE Trans Image Process 21(4):2035–2046
Hu P, Wu F, Peng J, Bao Y, Chen F, Kong D (2017) Automatic abdominal multi-organ segmentation using deep convolutional neural network and time-implicit level sets. Int J Comput Assist Radiol Surg 12(3):399–411
Shin H-C, Orton MR, Collins DJ, Doran SJ, Leach MO (2013) Stacked autoencoders for unsupervised feature learning and multiple organ detection in a pilot study using 4D patient data. IEEE Trans Pattern Anal Mach Intell 35(8):1930–1943
Wolz R, Chu C, Misawa K, Fujiwara M, Mori K, Rueckert D (2013) Automated abdominal multi-organ segmentation with subject-specific atlas generation. IEEE Trans Med Imaging 32(9):1723–1730
Zikic D, Glocker B, Criminisi A (2014) Encoding atlases by randomized classification forests for efficient multi-atlas label propagation. Med Image Anal 18(8):1262–1273
Oliveira B, Queiros S, Morais P, Torres HR, Gomes-Fonseca J, Fonseca JC, Vilaca JL (2018) A novel multi-atlas strategy with dense deformation field reconstruction for abdominal and thoracic multi-organ segmentation from computed tomography. Med Image Anal 45:108–120
Wang H, Zhang N, Huo L, Zhang B (2017) Evaluation of different atlas selection strategies for multi-atlas segmentation of low-dose computed tomographic images of whole-body positron emission tomography/computed tomography. Dig Med 3(4):186–192
Bagci U, Udupa JK, Mendhiratta N, Foster B, Xu Z, Yao J, Chen X, Mollura DJ (2013) Joint segmentation of anatomical and functional images: applications in quantification of lesions from PET, PET-CT, MRI-PET, and MRI-PET-CT images. Med Image Anal 17(8):929–945
Ballangan C, Wang X, Feng D (2011) Lung tumor delineation in PET-CT images based on a new segmentation energy. In: Nuclear science symposium and medical imaging conference, pp 3202–3205
Cui H, Wang X, Lin W, Zhou J, Eberl S, Feng D, Fulham M (2016) Primary lung tumor segmentation from PET–CT volumes with spatial–topological constraint. Int J Comput Assist Radiol Surg 11(1):1–11
Cui H, Wang X, Zhou J, Eberl S, Feng D, Fulham M (2015) Improved segmentation accuracy for thoracic PET-CT in patients with NSCLC using a multi-graph model (MGM). J Nucl Med 56(Suppl 3):2527
Cui H, Wang X, Zhou J, Eberl S, Yin Y, Feng D, Fulham M (2015) Topology polymorphism graph for lung tumor segmentation in PET-CT images. Phys Med Biol 60(12):4893–4914
Ju W, Xiang D, Zhang B, Wang L, Kopriva I, Chen X (2015) Random walk and graph cut for co-segmentation of Lung tumor on PET-CT images. IEEE Trans Image Process 24(12):5854–5867
Markel D, Caldwell C, Alasti H, Soliman H, Ung Y, Lee J, Sun A (2013) Automatic Segmentation of Lung Carcinoma Using 3D Texture Features in 18-FDG PET/CT. Int J Mol Imag 2013(980769):1–13
Song Q, Bai J, Han D, Bhatia S, Sun W, Rockey W, Bayouth JE, Buatti JM, Wu X (2013) Optimal co-segmentation of tumor in PET-CT images with context information. IEEE Trans Med Imaging 32(9):1685–1697
Xiang D, Chen X (2016) Automatic co-segmentation of lung tumor based on random forest in PET-CT images. In: Medical imaging 2016: image processing, 2016, vol 9784, pp 97842W1–97842W7
Bi L, Kim J, Wen L, Feng DD (2012) Automatic descending aorta segmentation in whole-body PET-CT studies for PERCIST-based thresholding. In: International conference on digital image computing techniques and applications, pp 1–6
Wang J, Xia Y, Feng DD (2012) Differential evolution based variational bayes inference for brain PET-CT image segmentation. In: International conference on digital image computing techniques and applications, pp 330–334
Xia AY, Wen L, Eberl S, Fulham M, Feng D (2009) Segmentation of brain PET-CT images based on adaptive use of complementary information. Proc SPIE Int Soc Opt Eng 7259:72593A1–72593A8
Xia Y, Eberl S, Feng D (2010) Dual-modality 3D brain PET-CT image segmentation based on probabilistic brain atlas and classification fusion. In: IEEE international conference on image processing, pp 2557–2560
Xia Y, Eberl S, Wen L, Fulham M, Feng DD (2012) Dual-modality brain PET-CT image segmentation based on adaptive use of functional and anatomical information. Comput Med Imag Gr 36(1):47–53
Xia Y, Wang J, Eberl S, Fulham M, Feng DD (2011) Brain tissue segmentation in PET-CT images using probabilistic atlas and variational Bayes inference. In: International conference of the IEEE engineering in medicine & biology society, pp 7969–7972
Xia Y, Wen L, Eberl S, Fulham M (2008) Segmentation of dual modality brain PET/CT images using the MAP-MRF model. In: 2008 IEEE workshop on multimedia signal processing, pp 107–110
Iglesias JE, Sabuncu MR (2014) Multi-atlas segmentation of biomedical images: a survey. Med Image Anal 24(1):205–219
Aljabar P, Heckemann RA, Hammers A, Hajnal JV, Rueckert D (2009) Multi-atlas based segmentation of brain images: atlas selection and its effect on accuracy. Neuroimage 46(3):726–738
Aljabar P, Heckemann R, Hammers A, Hajnal JV, Rueckert D (2007) Classifier selection strategies for label fusion using large atlas databases. Med Image Comput Comput Assist Interv 10(Pt 1):523–531
Aribisala BS, Cox SR, Ferguson KJ, Macpherson SE, Maclullich AM, Royle NA, Valdés Hernández MC, Bastin ME, Deary IJ, Wardlaw JM (2013) Assessing the performance of atlas-based prefrontal brain parcellation in an ageing cohort. J Comput Assist Tomogr 37(2):257–264
Rohlfing T, Brandt R, Menzel R, Maurer MC Jr (2004) Evaluation of atlas selection strategies for atlas-based image segmentation with application to confocal microscopy images of bee brains. Neuroimage 21(4):1428–1442
Heckemann RA, Hajnal JV, Aljabar P, Rueckert D, Hammers A (2006) Automatic anatomical brain MRI segmentation combining label propagation and decision fusion. Neuroimage 33(1):115–126
Wan J, Carass A, Resnick SM, Prince JL (2008) Automated reliable labeling of the cortical surface. Proc IEEE Int Symp Biomed Imaging 6:440–443
Beg MF, Miller MI, Trouvé A, Younes L (2005) Computing large deformation metric mappings via geodesic flows of diffeomorphisms. Int J Comput Vision 61(2):139–157
Vercauteren T, Pennec X, Perchant A, Ayache N (2009) Diffeomorphic demons: efficient non-parametric image registration. Neuroimage 45(1):S61–S72
Warfield SK, Zou KH, Wells WM (2004) Simultaneous truth and performance level estimation (STAPLE): an algorithm for the validation of image segmentation. IEEE Trans Med Imaging 23(7):903–921
Artaechevarria X, Munoz-Barrutia A, Ortiz-De-Solorzano C (2009) Combination strategies in multi-atlas image segmentation: application to brain MR data. IEEE Trans Med Imaging 28(8):1266–1277
Avants BB, Tustison NJ, Song G, Gee JC (2009) Ants: Open-source tools for normalization and neuroanatomy. Transac Med Imagins Penn Image Comput Sci Lab
Commowick O, Warfield SK (2009) A continuous STAPLE for scalar, vector, and tensor images: an application to DTI analysis. IEEE Trans Med Imag 28(6):838–846
Rajon DA, Bolch WE (2003) Marching cube algorithm: review and trilinear interpolation adaptation for image-based dosimetric models. Comput Med Imag Gr 27(5):411–435
Chui H, Rangarajan A (2000) A new algorithm for non-rigid point matching. In: Proceedings IEEE conference on computer vision and pattern recognition, pp 44–51
Glaister J, Carass A, Pham DL, Butman JA, Prince JL (2017) Falx cerebri segmentation via multi-atlas boundary fusion. In: International conference on medical image computing and computer-assisted intervention pp 92–99
Gardner MJ, Altman DG (1986) Confidence intervals rather than P values: estimation rather than hypothesis testing. BMJ 292(6522):746–750
Kohlberger T, Sofka M, Zhang J, Birkbeck N, Wetzl J, Kaftan J, Declerck J, Zhou SK (2011) Automatic multi-organ segmentation using learning-based segmentation and level set optimization. Med Image Comput Comput Assist Interv 14(Pt 3):338–345
Tong T, Wolz R, Wang Z, Gao Q, Misawa K, Fujiwara M, Mori K, Hajnal JV, Rueckert D (2015) Discriminative dictionary learning for abdominal multi-organ segmentation. Med Image Anal 23(1):92–104
Jimenez-del-Toro O, Muller H, Krenn M, Gruenberg K, Taha AA, Winterstein M, Eggel I, Foncubierta-Rodriguez A, Goksel O, Jakab A, Kontokotsios G, Langs G, Menze BH, Fernandez TS, Schaer R, Walleyo A, Weber MA, Cid YD, Gass T, Heinrich M, Jia FC, Kahl F, Kechichian R, Mai D, Spanier AB, Vincent G, Wang CL, Wyeth D, Hanbury A (2016) Cloud-based evaluation of anatomical structure segmentation and landmark detection algorithms: VISCERAL anatomy benchmarks. IEEE Trans Med Imag 35(11):2459–2475
Acknowledgements
This research is supported by the youth program of National Natural Science Fund of China No. 81401475, the general program of National Natural Science Fund of China No. 61571076, the general program of Liaoning Science and Technology Project No. 2015020040, the Since and Technology Star Project Fund of Dalian City No. 2016RQ019, the Xinghai Scholar Cultivating Funding of Dalian University of Technology (No. DUT14RC(3)066), the cultivating program of Major National Natural Science Fund of China No. 91546123, the National Key Research and Development Program No. 2016YFC0103101, 2016YFC0103102 and 2016YFC0106402, and the Fundamental Research Funds for Central Universities No. DUT15LN02 and No. DUT16RC(3)099.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Human and animal rights
This article does not contain any studies with animals performed by any of the authors.
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Declaration of Helsinki and its later amendments or comparable ethical standards.
Informed consent
Statement of informed consent is not applicable since the manuscript does not contain any participants’ data.
Rights and permissions
About this article
Cite this article
Wang, H., Zhang, N., Huo, L. et al. Dual-modality multi-atlas segmentation of torso organs from [18F]FDG-PET/CT images. Int J CARS 14, 473–482 (2019). https://doi.org/10.1007/s11548-018-1879-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11548-018-1879-3