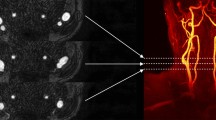
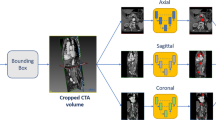
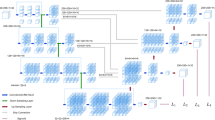
Abstract
Purpose
Carotid artery atherosclerotic stenosis accounts for 18–25% of ischemic stroke. In the evaluation of carotid atherosclerotic lesions, the automatic, accurate and rapid segmentation of the carotid artery is a priority issue that needs to be addressed urgently. However, the carotid artery area occupies a small target in computed tomography angiography (CTA) images, which affect the segmentation accuracy.
Methods
We proposed a coarse-to-fine segmentation pipeline with the Multiplanar D-SEA UNet to achieve fully automatic carotid artery segmentation on the entire 3D CTA images, and compared with other four neural networks (3D-UNet, RA-UNet, Isensee-UNet, Multiplanar-UNet) by assessing Dice, Jaccard similarity coefficient, sensitivity, area under the curve and average hausdorff distance.
Results
Our proposed method can achieve a mean Dice score of 91.51% on the 68 neck CTA scans from Beijing Hospital, which remarkably outperforms state-of-the-art 3D image segmentation methods. And the C2F segmentation pipeline can effectively improve segmentation accuracy while avoiding resolution loss.
Conclusion
The proposed segmentation method can realize the fully automatic segmentation of the carotid artery and has robust performance with segmentation accuracy, which can be applied into plaque exfoliation and interventional surgery services. In addition, our method is easy to extend to other medical segmentation tasks with appropriate parameter settings.






Similar content being viewed by others
Abbreviations
- CTA:
-
Computed tomography angiography
- CNN:
-
Convolutional neural network
- DSA:
-
Digital subtraction angiography
- ECAD:
-
Extracranial atherosclerotic disease
- C2F:
-
Coarse-to-fine
- Sen:
-
Sensitivity
- AUC:
-
Area under the curve
- AVD:
-
Average Hausdorff distance
References
Collaborators G2 (2018) Global, regional, and national disability-adjusted life-years (DALYs) for 359 diseases and injuries and healthy life expectancy (HALE) for 195 countries and territories, 1990–2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet (London, England) 392:1859–1922
Brott T, Halperin J, Abbara S, Bacharach J, Barr J, Bush R, Cates CU, Creager M, Fowler SB, Friday G, Hertzberg V, McIff EB, Moore W, Panagos P, Riles T, Rosenwasser R, Taylor A (2011) 2011 ASA/ACCF/AHA/AANN/AANS/ACR/ASNR/CNS/SAIP/SCAI/SIR/SNIS/SVM/SVS guideline on the management of patients with extracranial carotid and vertebral artery disease: executive summary. Stroke 42(8):e420–e463
Ooi Y, Gonzalez N (2015) Management of extracranial carotid artery disease. Cardiol Clin 33(1):1–35
Saba L, Saam T, Jäger H, Yuan C, Wintermark M (2019) Imaging biomarkers of vulnerable carotid plaques for stroke risk prediction and their potential clinical implications. Lancet Neurol 18:559–572
Saxena A, Ng E, Lim S (2019) Imaging modalities to diagnose carotid artery stenosis: progress and prospect. BioMed Eng Online 18:1–23
Beare R, Chong W, Ren M, Das G, Srikanth V, Phan T (2010) Segmentation of carotid arteries in CTA images. Int Conf Digital Image Comput Tech Appl 2010:69–74
Santos FL, Joutsen A, Terada M, Salenius J, Eskola H (2014) A semi-automatic segmentation method for the structural analysis of carotid atherosclerotic plaques by computed tomography angiography. J Atheroscler Thromb 21(9):930–940
Bozkurt F, Köse C, Sarı A (2018) An inverse approach for automatic segmentation of carotid and vertebral arteries in CTA. Expert Syst Appl 93:358–375
Tang H, Walsum TV, Hameeteman R, Shahzad R, Vliet LV, Niessen W (2013) Lumen segmentation and stenosis quantification of atherosclerotic carotid arteries in CTA utilizing a centerline intensity prior. Med Phys 40(5):051721
Zhou R, Fenster A, Xia Y, Spence J, Ding M (2019) Deep learning-based carotid media-adventitia and lumen-intima boundary segmentation from three-dimensional ultrasound images. Med Phys 46:3180–3193
Moreira AS (2019) Carotid lumen segmentation using a neural network approach. Dissertation, University of Porto
Pan XY (2019) Carotid bifurcation segmentation based on the deep neural network UNet. Dissertation, Zhejiang University
Hameeteman R, Zuluaga MA, Freiman M, Joskowicz L, Cuisenaire O, Florez-Valencia L, Gülsün M, Krissian K, Mille J, Wong WC, Orkisz M, Tek H, Hoyos MH, Benmansour F, Chung AC, Rozie S, Gils MV, Borne LV, Sosna J, Berman P, Cohen N, Douek P, Sánchez I, Aissat M, Schaap M, Metz C, Krestin G, Lugt A, Niessen W, Walsum TV (2011) Evaluation framework for carotid bifurcation lumen segmentation and stenosis grading. Med Image Anal 15(4):477–488
Ziegler M, Alfraeus J, Bustamante M, Good E, Engvall J, de Muinck E, Dyverfeldt P (2021) Automated segmentation of the individual branches of the carotid arteries in contrast-enhanced MR angiography using DeepMedic. BMC Med Imag 21(1):1–10
Perslev M, Dam E, Pai A, Igel C (2019) One network to segment them all: a general, lightweight system for accurate 3D medical image segmentation. In: MICCAI, pp 30–38
Ronneberger O, Fischer P, Brox T (2015) U-Net: convolutional networks for biomedical image segmentation. In: Navab N, Hornegger J, Wells W, Frangi A (eds) Medical image computing and computer-assisted intervention – MICCAI 2015. MICCAI 2015. Lecture notes in computer science, vol 9351, Springer, pp 234–241
Isensee F, Kickingereder P, Wick W, Bendszus M, Maier-Hein KH (2018) Brain tumor segmentation and radiomics survival prediction: contribution to the BRATS 2017 challenge. In: Crimi A, Bakas S, Kuijf H, Menze B, Reyes M (eds) Brainlesion: glioma, multiple sclerosis, stroke and traumatic brain injuries. BrainLes 2017. Lecture notes in computer science, vol 10670, Springer, pp 287–297
Roy AG, Navab N, Wachinger C (2019) Recalibrating fully convolutional networks with spatial and channel “squeeze and excitation” blocks. IEEE Trans Med Imag 38:540–549
Oktay O, Schlemper J, Folgoc LL, Lee MJ, Heinrich M, Misawa K, Mori K, McDonagh SG, Hammerla N, Kainz B, Glocker B, Rueckert D (2018). Attention U-Net: learning where to look for the pancreas. http://arxiv.org/abs/1804.03999.
Castro E, Cardoso JS, Pereira JC (2018) Elastic deformations for data augmentation in breast cancer mass detection. IEEE EMBS Int Conf Biomed Health Inform (BHI) 2018:230–234
Taha AA, Hanbury A (2015) Metrics for evaluating 3D medical image segmentation: analysis, selection, and tool. BMC Med Imag 15(1):29
Powers D (2011) Evaluation: from precision, recall and F-factor to ROC, informedness, markedness correlation. Mach Learn Technol 2:37–63
Çiçek Ö, Abdulkadir A, Lienkamp SS, Brox T, Ronneberger O (2016) 3D U-Net: learning dense volumetric segmentation from sparse annotation. In: Ourselin S, Joskowicz L, Sabuncu M, Unal G, Wells W (eds) Medical image computing and computer-assisted intervention – MICCAI 2016. MICCAI 2016. Lecture notes in computer science, vol 9901, Springer, pp 424–432
Jin Q, Meng Z, Sun C, Wei L, Su R (2020) RA-UNet: a hybrid deep attention-aware network to extract liver and tumor in CT scans. Front Bioeng Biotechnol 8:1471
Kou C, Li W, Liang W, Yu Z, Hao J (2019) Microaneurysms segmentation with a U-Net based on recurrent residual convolutional neural network. J Med Imag 6:025008–025008
Funding
This work was funded by the Capital’s Funds for Health Improvement and Research (2020-4-4053), the National Natural Science Foundation of China (KKA309004533), the Enterprise funded projects from Fudan University (20275), the Independent Research fund of Key Laboratory of Industrial Dust Prevention and Control & Occupational Health and Safety, Ministry of Education (Anhui University of Science and Technology) (EK20201003) and the Shandong Key Laboratory of Intelligent Buildings Technology (SDIBT202006).
Author information
Authors and Affiliations
Contributions
JW collected and labeled the medical images. YY and RY conducted the experiments, wrote the draft. HW and DG participated in data and material review. JL and ZY participated in experiment design, reviewed and edited the manuscript.
Corresponding authors
Ethics declarations
Consent to participate
Agree.
Consent for publication
Agree
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
For the retrospective studies formal consent is not required.
Informed consent
Informed consent was obtained from all individual participants included in the study.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Wang, J., Yu, Y., Yan, R. et al. Coarse-to-fine multiplanar D-SEA UNet for automatic 3D carotid segmentation in CTA images. Int J CARS 16, 1727–1736 (2021). https://doi.org/10.1007/s11548-021-02471-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11548-021-02471-5