Abstract
Purpose
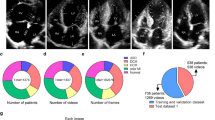
Carpentier’s functional classification is a guide to explain the types of mitral valve regurgitation based on morphological features. There are four types of pathological morphologies, regardless of the presence or absence of mitral regurgitation: Type I, normal; Type II, mitral valve prolapse; Type IIIa, mitral valve stenosis; and Type IIIb, restricted mitral leaflet motion. The aim of this study was to automatically classify mitral valves using echocardiographic images.
Methods
In our procedure, after the classification of apical 4-chamber (A4C) and parasternal long-axis (PLA) views, we extracted the systolic/diastolic phase of the cardiac cycle by calculating the left ventricular area. Six typical pre-trained models were fine-tuned with a 4-class model for the PLA and a 3-class model for the A4C views. As an additional contribution, to provide explainability, we applied the Gradient-weighted Class Activation Mapping (Grad-CAM) algorithm to visualize areas of echocardiographic images where the different models generated a prediction.
Results
This approach conferred a proper understanding of where various networks “look” into echocardiographic images to predict the four types of pathological mitral valve morphologies. Considering the accuracy metric and Grad-CAM maps and by applying the Inception-ResNet-v2 architecture to classify Type II in the PLA view and ResNeXt50 architecture to classify the other three classes in the A4C view, we achieved an 80% rate of model accuracy in the test data set.
Conclusions
We suggest an explainable, fully automated, and rule-based procedure to classify the four types of mitral valve morphologies based on Carpentier’s functional classification using deep learning on transthoracic echocardiographic images. Our study results infer the feasibility of the use of deep learning models to prepare quick and precise assessments of mitral valve morphologies in echocardiograms. According to our knowledge, our study is the first one that provides a public data set regarding the Carpentier classification of MV pathologies.














Similar content being viewed by others
Data availability
The datasets generated during the current study are available from the corresponding author on reasonable request at https://github.com/medical-dataset/Mitral-Valve-Echocardiography.
Code availability
Code for data cleaning and analysis are available from the corresponding author on reasonable request.
References
Bonow RO, O’Gara PT, Adams DH, Badhwar V, Bavaria JE, Elmariah S, Hung JW, Lindenfeld J, Morris AA, Satpathy R (2020) 2020 Focused update of the 2017 ACC expert consensus decision pathway on the management of mitraláregurgitation: a report of the american college of cardiology solution set oversight committee. J Am Coll Cardiol 75(17):2236–2270. https://doi.org/10.1016/j.jacc.2020.02.005
Carpentier A (1983) Cardiac valve surgery—the “French correction.” J Thorac Cardiovasc Surg 86(3):323–337. https://doi.org/10.1016/S0022-5223(19)39144-5
Carpentier A, Adams DH, Filsoufi F (2011) Carpentier’s Reconstructive Valve Surgery E-Book. Elsevier Health Sciences
Fourcade A, Khonsari R (2019) Deep learning in medical image analysis: a third eye for doctors. J Stomatol Oral Maxillofac Surg 120(4):279–288. https://doi.org/10.1016/j.jormas.2019.06.002
Østvik A, Smistad E, Aase SA, Haugen BO, Lovstakken L (2019) Real-time standard view classification in transthoracic echocardiography using convolutional neural networks. Ultrasound Med Biol 45(2):374–384. https://doi.org/10.1016/j.ultrasmedbio.2018.07.024
Vafaeezadeh M, Behnam H, Hosseinsabet A, Gifani P (2021) A deep learning approach for the automatic recognition of prosthetic mitral valve in echocardiographic images. Comput Biol Med 133:104388–1
Ge R, Yang G, Chen Y, Luo L, Feng C, Ma H, Ren J, Li S (2019) K-net: Integrate left ventricle segmentation and direct quantification of paired echo sequence. IEEE Trans Med Imaging 39(5):1690–1702
Jafari MH, Girgis H, Van Woudenberg N, Liao Z, Rohling R, Gin K, Abolmaesumi P, Tsang T (2019) Automatic biplane left ventricular ejection fraction estimation with mobile point-of-care ultrasound using multi-task learning and adversarial training. Int J Comput Assist Radiol Surg 14(6):1027–1037. https://doi.org/10.1007/s11548-019-01954-w
Leclerc S, Smistad E, Pedrosa J, Østvik A, Cervenansky F, Espinosa F, Espeland T, Berg EAR, Jodoin P-M, Grenier T (2019) Deep learning for segmentation using an open large-scale dataset in 2D echocardiography. IEEE Trans Med Imaging 38(9):2198–2210
Smistad E, Østvik A, Salte IM, Melichova D, Nguyen TM, Haugaa K, Brunvand H, Edvardsen T, Leclerc S, Bernard O (2020) Real-time automatic ejection fraction and foreshortening detection using deep learning. IEEE Trans Ultrason Ferroelectr Freq Control 67(12):2595–2604. https://doi.org/10.1109/tuffc.2020.2981037
Kusunose K, Abe T, Haga A, Fukuda D, Yamada H, Harada M, Sata M (2020) A deep learning approach for assessment of regional wall motion abnormality from echocardiographic images. Cardiovasc Imaging 13(2):374–381
Sotaquira M, Pepi M, Fusini L, Maffessanti F, Lang RM, Caiani EG (2015) Semi-automated segmentation and quantification of mitral annulus and leaflets from transesophageal 3-D echocardiographic images. Ultrasound Med Biol 41(1):251–267
Pedrosa J, Queirós S, Vilaça J, Badano L, D'hooge J. Fully automatic assessment of mitral valve morphology from 3D transthoracic echocardiography. In: 2018 IEEE International ultrasonics symposium (IUS). IEEE; 2018. p. 1–6.
Andreassen BS, Veronesi F, Gerard O, Solberg AHS, Samset E (2019) Mitral annulus segmentation using deep learning in 3-D transesophageal echocardiography. IEEE J Biomed Health Inform 24(4):994–1003
Xie S, Girshick R, Dollár P, Tu Z, He K. Aggregated residual transformations for deep neural networks. In: Proceedings of the IEEE conference on computer vision and pattern recognition; 2017. p. 1492–1500.
Tan M, Le Q. Efficientnet: rethinking model scaling for convolutional neural networks. In: International conference on machine learning. PMLR; 2019. p. 6105–6114.
Darvishi S, Behnam H, Pouladian M, Samiei N (2013) Measuring left ventricular volumes in two-dimensional echocardiography image sequence using level-set method for automatic detection of end-diastole and end-systole frames. Res Cardiovasc Med 2(1):39
Tajbakhsh N, Shin JY, Gurudu SR, Hurst RT, Kendall CB, Gotway MB, Liang J (2016) Convolutional neural networks for medical image analysis: Full training or fine tuning? IEEE Trans Med Imaging 35(5):1299–1312
Deng J, Dong W, Socher R, Li L-J, Li K, Fei-Fei L. Imagenet: a large-scale hierarchical image database. In: 2009 IEEE conference on computer vision and pattern recognition. IEEE; 2009. p. 248–255.
Zoph B, Vasudevan V, Shlens J, Le QV. Learning transferable architectures for scalable image recognition. In: Proceedings of the IEEE conference on computer vision and pattern recognition; 2018. p. 8697–8710. https://doi.org/10.1109/CVPR.2018.00907.
Chollet, F. Xception: deep learning with depthwise separable convolutions. In: Proceedings of the IEEE conference on computer vision and pattern recognition; 2017. p. 1251–1258.
Szegedy C, Ioffe S, Vanhoucke V, Alemi A. Inception-v4, inception-resnet and the impact of residual connections on learning. In: Proceedings of the AAAI conference on artificial intelligence, vol. 1; 2017.
Selvaraju RR, Cogswell M, Das A, Vedantam R, Parikh D, Batra D. Grad-cam: visual explanations from deep networks via gradient-based localization. In: Proceedings of the IEEE international conference on computer vision; 2017. p. 618–626. https://doi.org/10.1109/ICCV.2017.74.
Funding
Not applicable.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors have no conflicts of interest to declare that are relevant to the content of this article.
Ethical approval
Ethical approval for this study was granted by the Ethics Committee of Tehran Heart Center, Iran.
Informed consent
Informed consent was obtained from all individual participants included in the study.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Vafaeezadeh, M., Behnam, H., Hosseinsabet, A. et al. Automatic morphological classification of mitral valve diseases in echocardiographic images based on explainable deep learning methods. Int J CARS 17, 413–425 (2022). https://doi.org/10.1007/s11548-021-02542-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11548-021-02542-7