Abstract
Purpose
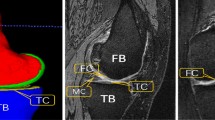
Accurate segmentation of articular cartilage from MR images is crucial for quantitative investigation of pathoanatomical conditions such as osteoarthritis (OA). Recently, deep learning-based methods have made significant progress in hard tissue segmentation. However, it remains a challenge to develop accurate methods for automatic segmentation of articular cartilage.
Methods
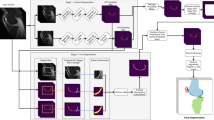
We propose a two-stage method for automatic segmentation of articular cartilage. At the first stage, nnU-Net is employed to get segmentation of both hard tissues and articular cartilage. Based on the initial segmentation, we compute distance maps as well as entropy maps, which encode the uncertainty information about the initial cartilage segmentation. At the second stage, both distance maps and entropy maps are concatenated to the original image. We then crop a sub-volume around the cartilage region based on the initial segmentation, which is used as the input to another nnU-Net for segmentation refinement.
Results
We designed and conducted comprehensive experiments on segmenting three different types of articular cartilage from two datasets, i.e., an in-house dataset consisting of 25 hip MR images and a publicly available dataset from Osteoarthritis Initiative (OAI). Our method achieved an average Dice similarity coefficient (DSC) of \(92.1\pm 0.99\%\) for the combined hip cartilage, \(89.8\pm 2.50\%\) for the femoral cartilage and \(86.4\pm 4.13\%\) for the tibial cartilage, respectively.
Conclusion
In summary, we developed a new approach for automatic segmentation of articular cartilage from MR images. Comprehensive experiments conducted on segmenting articular cartilage of the knee and hip joints demonstrated the efficacy of the present approach. Our method achieved equivalent or better results than the state-of-the-art methods.


Similar content being viewed by others
References
Ambellan F, Tack A, Ehlke M, Zachow S (2019) Automated segmentation of knee bone and cartilage combining statistical shape knowledge and convolutional neural networks: Data from the osteoarthritis initiative. Med Image Anal 52:109–118
Cheng Y, Guo C, Wang Y, Bai J, Tamura S (2012) Accuracy limits for the thickness measurement of the hip joint cartilage in 3-d MR images: simulation and validation. IEEE Trans Biomed Eng 60(2):517–533
Çiçek Ö, Abdulkadir A, Lienkamp SS, Brox T, Ronneberger O (2016) 3d u-net: learning dense volumetric segmentation from sparse annotation. In: International conference on medical image computing and computer-assisted intervention. pp. 424–432. Springer
Drozdzal M, Vorontsov E, Chartrand G, Kadoury S, Pal C (2016) The importance of skip connections in biomedical image segmentation. In Deep learning and data labeling for medical applications, pp 179–187. Springer
Fripp J, Crozier S, Warfield SK, Ourselin S (2007) Automatic segmentation of the bone and extraction of the bone-cartilage interface from magnetic resonance images of the knee. Phys Med Biol 52(6):1617
Heimann T, Morrison BJ, Styner MA, Niethammer M, Warfield S (2010) Segmentation of knee images: a grand challenge. In Proceedings of MICCAI Workshop on Medical Image Analysis for the Clinic. pp 207–214. Beijing, China
Isensee F, Jaeger PF, Kohl SA, Petersen J, Maier-Hein KH (2021) nnu-net: a self-configuring method for deep learning-based biomedical image segmentation. Nat Methods 18(2):203–211
Lee JG, Gumus S, Moon CH, Kwoh CK, Bae KT (2014) Fully automated segmentation of cartilage from the MR images of knee using a multi-atlas and local structural analysis method. Med Phys 41(9):092303
Liu F, Zhou Z, Jang H, Samsonov A, Zhao G, Kijowski R (2018) Deep convolutional neural network and 3d deformable approach for tissue segmentation in musculoskeletal magnetic resonance imaging. Magn Reson Med 79(4):2379–2391
Paszke A, Gross S, Massa F, Lerer A, Bradbury J, Chanan G, Killeen T, Lin Z, Gimelshein N, Antiga L, Desmaison A, Köpf A, Yang E, DeVito Z, Raison M, Tejani A, Chilamkurthy S, Steiner B, Fang L, Bai J, Chintala S (2019) Pytorch: An imperative style, high-performance deep learning library. arXiv preprint arXiv:1912.01703
Pedoia V, Majumdar S, Link TM (2016) Segmentation of joint and musculoskeletal tissue in the study of arthritis. Magn Reson Mater Phys Biol Med 29(2):207–221
Prasoon A, Petersen K, Igel C, Lauze F, Dam E, Nielsen M (2013) Deep feature learning for knee cartilage segmentation using a triplanar convolutional neural network. In International conference on medical image computing and computer-assisted intervention. pp 246–253. Springer
Schmaranzer F, Helfenstein R, Zeng G, Lerch TD, Novais EN, Wylie JD, Kim YJ, Siebenrock KA, Tannast M, Zheng G (2019) Automatic MRI-based three-dimensional models of hip cartilage provide improved morphologic and biochemical analysis. Clinical Orthop Relat Res 477(5):1036
Shan L, Zach C, Charles C, Niethammer M (2014) Automatic atlas-based three-label cartilage segmentation from MR knee images. Med Image Anal 18(7):1233–1246
Siversson C, Akhondi-Asl A, Bixby S, Kim YJ, Warfield SK (2014) Three-dimensional hip cartilage quality assessment of morphology and Dgemric by planar maps and automated segmentation. Osteoarth Cartil 22(10):1511–1515
Xia Y, Chandra SS, Engstrom C, Strudwick MW, Crozier S, Fripp J (2014) Automatic hip cartilage segmentation from 3d MR images using arc-weighted graph searching. Phys Med Biol 59(23):7245
Acknowledgements
The work was partially supported by the Key Program of the Medical Engineering Interdisciplinary Research Fund of Shanghai Jiaotong University via project YG2019ZDA22 and YG2019ZDB09, and by the Natural Science Foundation of China via project U20A20199. Osteoarthritis Initiative is a public-private partnership comprised of five contracts (N01-AR-2-2258; N01-AR-2-2259; N01-AR-2-2260; N01-AR-2-2261; N01-AR-2-2262) funded by the National Institutes of Health, a branch of the Department of Health and Human Services, and conducted by the OAI Study Investigators. Private funding partners include Merck Research Laboratories; Novartis Pharmaceuticals Corporation, GlaxoSmithKline; and Pfizer, Inc. Private sector funding for the OAI is managed by the Foundation for the National Institutes of Health. This manuscript was prepared using an OAI public use dataset and does not necessarily reflect the opinions or views of the OAI investigators, the NIH, or the private funding partners.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Informed consent
Informed consent was obtained from all individuals included in the study.
Ethics approval
Institutional ethics approval was obtained for the use of clinical data in this study.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Li, Z., Chen, K., Liu, P. et al. Entropy and distance maps-guided segmentation of articular cartilage: data from the Osteoarthritis Initiative. Int J CARS 17, 553–560 (2022). https://doi.org/10.1007/s11548-021-02555-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11548-021-02555-2