Abstract
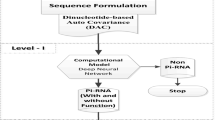
The significant role of long non-coding RNAs (lncRNAs) in various cellular functions, such as gene imprinting, immune response, embryonic pluripotency, tumorogenesis, and genetic regulations, has been widely studied and reported in recent years. Several experimental and computational methods involving genome-wide search and screenings of ncRNAs are being proposed utilizing sequence features-length, occurrence, and composition of bases with various limitations. The proposed classifier, Deep Neural Network (DNN) is fast and an accurate alternative for the identification of lncRNAs as compared to other existing classifiers. The information content stored in k-mer pattern has been used as a sole feature for the DNN classifier using manually annotated training datasets from LNCipedia and RefSeq database, obtaining accuracy of 98.07 %, sensitivity of 98.98 %, and specificity of 97.19 %, respectively, on test dataset. The k-mer information content generated on the basis of Shannon entropy function has resulted in improved classifier accuracy. This classification framework was also tested on known human genome dataset, and the framework has successfully identified known lncRNAs with 99 % accuracy rate. The said algorithm has been implemented as a web prediction tool, which is available on server interface http://bioserver.iiita.ac.in/deeplnc.





Similar content being viewed by others
References
Akhter S, Bailey B, Salamon P, Aziz RK, Edwards R (2013) Applying Shannonʼs information theory to bacterial and phage genomes and metagenomes. Sci Reports 3:1033
Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W et al (1997) Gapped BLAST and PSI BLAST: A new generation of protein database search programs. Nucleic Acids Res 25(17):3389–3402
Amaral PP, Clark MB, Gascoigne DK, Dinger ME, Mattick JS (2011) LncRNAdb: a reference database for long noncoding RNAs. Nucleic Acids Res 39(Database issue):D146–D151
An J, Lai J, Lehman ML, Nelson C (2013) MiRDeep*: an integrated application tool for miRNA identification from RNA sequencing data. Nucleic Acids Res 41(2):727–737
Babak T, Blencowe BJ, Hughes TR (2005) A systematic search for new mammalian noncoding RNAs indicates little conserved intergenic transcription. BMC Genom 6:104
Badger JH, Olsen GJ (1999) CRITICA: coding region identification tool invoking comparative analysis. Mol Biol Evol 16(4):512–524
Baker M (2011) Long noncoding RNAs: the search for function. Nat Methods 8(5):379–383
Berg JM, Tymoczko JL, Stryer L (2002) Biochemistry. W H Freeman, New York
Bhartiya D, Pal K, Ghosh S, Kapoor S, Jalali S, Panwar B et al (2013) LncRNome: a comprehensive knowledgebase of human long noncoding RNAs. Database (Oxford) 2013:bat034. doi:10.1093/database/bat034
Bottou L (2010) Large-scale machine learning with stochastic gradient descent. In: Proceedings of COMPSTATʼ10, pp 177–186
Chen X, Gui Y (2013) Novel human lncRNA-disease association inference based on lncRNA expression profiles. Bioinformatics 29(20):2617–2624
Chen G, Wang Z, Wang D, Qiu C, Liu M, Chen X et al (2013) LncRNADisease: a database for long-non-coding RNA-associated diseases. Nucleic Acids Res 41(Database issue):D983–D986
Clement C, Hill JM, Dua P, Culicchia F, Lukiw WJ (2016) Analysis of RNA from Alzheimer’s Disease Post-mortem Brain Tissues. Mol Neurobiol 53(2):1322–1328. doi:10.1007/s12035-015-9105-6
Chu C, Qu K, Zhong FL, Artandi SE, Chang HY (2011) Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Mol Cell 44(4):667–678
Clamp M, Fry B, Kamal M, Xie X, Cuff J, Lin MF et al (2007) Distinguishing protein-coding and noncoding genes in the human genome. Proc Natl Acad Sci USA 104(49):19428–19433
Coronnello C, Hartmaier R, Arora A, Huleihel L, Pandit KV, Bais AS et al (2012) Novel modeling of combinatorial miRNA Targeting identifies SNP with potential role in bone density. PLoS Comput Biol 8(12):e1002830 (Print)
Dinger ME, Pang KC, Mercer TR, Crowe ML, Grimmond SM, Mattick JS (2009) NRED: a database of long noncoding RNA expression. Nucleic Acids Res 37(Suppl. 1):D122–D126
Duchi J, Hazan E, Singer Y (2011) Adaptive subgradient methods for online learning and stochastic optimization. J Mach Learn Res 12:2121–2159
Engelen S, Tahi F (2010) Tfold: efficient in silico prediction of non-coding RNA secondary structures. Nucleic Acids Res 38(7):2453–2466
Furuno M, Pang KC, Ninomiya N, Fukuda S, Frith MC, Bult C, Kai C, Kawai J, Carninci P, Hayashizaki Y, Mattick JS, Suzuki H (2006) Clusters of internally primed transcripts reveal novel long noncoding RNAs. PLoS Genet 2(4):537–553
Gibb EA, Vucic EA, Enfield KSS, Stewart GL, Lonergan KM, Kennett JY et al (2011) Human cancer long non-coding RNA transcriptomes. PLoS One 6(10):e25915 (Print)
Goff LA, Rinn J (2015) Linking RNA biology to lncRNAs. Genome Res. Cold Spring Harbor Laboratory Press 25(10):1456–1465
Granovskaia MV, Jensen LJ, Ritchie ME, Toedling J, Ning Y, Bork P, Wolfgang H, Steinmetz LM (2010) High-resolution transcription atlas of the mitotic cell cycle in budding yeast. Genome Biol 11(3):R24
Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D et al (2009) Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 458(7235):223–227
Harries LW (2012) Long non-coding RNAs and human disease. Biochem Soc Trans 40(4):902–906
Haubold B, Pierstorff N, Moller F, Wiehe T (2005) Genome comparison without alignment using shortest unique substrings. BMC Bioinform 6(1):123
Hu W, Yuan B, Flygare J, Lodish HF (2011) Long noncoding RNA-mediated anti-apoptotic activity in murine erythroid terminal differentiation. Genes Dev 25(24):2573–2578
Huang W, Long N, Khatib H (2012) Genome-wide identification and initial characterization of bovine long non-coding RNAs from EST data. Anim Gene 43(6):674–682
Hüttenhofer A, Schattner P, Polacek N (2005) Non-coding RNAs: hope or hype? Trends Genet 21:289–297
Jiang Q, Wang J, Wang Y, Ma R, Wu X, Li Y (2014) TF2LncRNA: identifying common transcription factors for a list of lncRNA genes from ChIP-seq data. BioMed Res Int 2014:317642. doi:10.1155/2014/317642
Jin J, Liu J, Wang H, Wong L, Chua NH (2013) PLncDB: plant long non-coding RNA database. Bioinformatics 29(8):1068–1071
Krizhevsky A, Sutskever I, Hinton GE (2012) Image net classification with deep convolutional neural networks. Advances in neural information processing systems, pp 1–9
Kung JTY, Colognori D, Lee JT (2013) Long noncoding RNAs: past, present, and future. Genetics 193(3):651–669
Lasda E, Roy P (2014) Circular RNAs: diversity of form and function. RNA (New York, N.Y.) 20(12):1829–1842
Lee H, Grosse R, Ranganath R, Ng AY (2009) Convolutional deep belief networks for scalable unsupervised learning of hierarchical representations. In: Proceedings of the 26th Annual International Conference on Machine Learning ICML 09, pp 1–8
Lertampaiporn S, Thammarongtham C, Nukoolkit C, Kaewkamnerdpong B, Ruengjitchatchawalya M (2014) Identification of non-coding RNAs with a new composite feature in the Hybrid Random Forest Ensemble algorithm. Nucleic Acids Res 42(11):e93. doi:10.1093/nar/gku325
Li A, Zhang J, Zhou Z (2014) PLEK: a tool for predicting long non-coding RNAs and messenger RNAs based on an improved k-mer scheme. BMC Bioinform 15:311
Liao Q, Xiao H, Bu D, Xie C, Miao R, Luo H et al (2011) NcFANs: a web server for functional annotation of long non-coding RNAs. Nucleic Acids Res 39(Suppl):2
Lin MF, Jungreis I, Kellis M (2011) PhyloCSF: a comparative genomics method to distinguish protein coding and non-coding regions. Bioinformatics 27(13):i275–i282
Liu J, Gough J, Rost B (2006) Distinguishing protein-coding from non-coding RNAs through support vector machines. PLoS Genet 2:529–536
Ma H, Hao Y, Dong X, Gong Q, Chen J, Zhang J, Tian W (2012) Molecular mechanisms and function prediction of long noncoding RNA. Sci World J 2012(1):541786
Marques AC, Hughes J, Graham B, Kowalczyk MS, Higgs DR, Ponting CP (2013) Chromatin signatures at transcriptional start sites separate two equally populated yet distinct classes of intergenic long noncoding RNAs. Genome Biol 14(11):R131
Morris KV, Mattick JS (2014) The rise of regulatory RNA. Nat Rev Genet 15(6):423–437
Nesterov Y (2007) Gradient methods for minimizing composite objective function. Core discussion paper. ReCALL 76.2007076 (2007): 2007/76
Niazi F, Valadkhan S (2012) Computational analysis of functional long noncoding RNAs reveals lack of peptide-coding capacity and parallels with 3ʼ UTRs. RNA 18(4):825–843
Nie L, Wu HJ, Hsu JM, Chang SS, LaBaff AM, Li CW, Wang Y, Hsu JL, Hung MC (2012) Long non-coding RNAs: versatile master regulators of gene expression and crucial players in cancer. Am J Transl Res 4(2):127–150
Paraskevopoulou MD, Georgakilas G, Kostoulas N, Reczko M, Maragkakis M, Dalamagas TM, Hatzigeorgiou AG (2013) DIANA-LncBase: experimentally verified and computationally predicted microRNA targets on long non-coding RNAs. Nucleic Acids Res 41(D1):D239–D245
Park C, Yu N, Choi I, Kim W, Lee S (2014) lncRNAtor: a comprehensive resource for functional investigation of long non-coding RNAs. Bioinformatics 30(17):2480–2485
Pasmant E, Laurendeau I, Héron D, Vidaud M, Vidaud D, Bièche I (2007) Characterization of a germ-line deletion, including the entire INK4/ARF locus, in a melanoma-neural system tumor family: identification of ANRIL, an antisense noncoding RNA whose expression coclusters with ARF. Cancer Res 67(8):3963–3969
Ponting CP, Oliver PL, Reik W (2009) Evolution and functions of long noncoding RNAs. Cell 136(4):629–641
Prensner JR, Chinnaiyan AM (2011) The emergence of lncRNAs in cancer biology. Cancer Discov 1(5):391–407
Qinghua J, Rui M, Jixuan W, Xiaoliang W, Shuilin J, Jiajie P, Tan R, Zhang T, Li Y, Wang Y (2015) LncRNA2Function: a comprehensive resource for functional investigation of human lncRNAs based on RNA-seq data. BMC Genom 16(3):S2
Rè M, Pesole G, Horner DS (2009) Accurate discrimination of conserved coding and non-coding regions through multiple indicators of evolutionary dynamics. BMC Bioinformatics 10:282. doi:10.1186/1471-2105-10-282
Rinn JL (2014) LncRNAs: linking RNA to chromatin. Cold Spring Harb Perspect Biol 6(8). pii: a018614. doi:10.1101/cshperspect.a018614
Sacco LDA, Baldassarre A, Masotti A (2012) Bioinformatics tools and novel challenges in long non-coding RNAs (lncRNAs) functional analysis. Int J Mol Sci 13(1):97–114
Sales G, Coppe A, Bisognin A, Biasiolo M, Bortoluzzi S, Romualdi C (2010) Magia, a web-based tool for miRNA and genes integrated analysis. Nucleic Acids Res 38(2). (Print)
Simon MD (2013) Capture hybridization analysis of RNA targets (CHART). Curr Protoc Mol Biol. doi:10.1002/0471142727.mb2125s101
Singh DK, Prasanth KV (2013) Functional insights into the role of nuclear-retained long noncoding RNAs in gene expression control in mammalian cells. Chromosome Res Int J Mole Supramole Evolut Aspects Chromosome Biol 21(6–7):695–711
Sun L, Zhang Z, Bailey TL, Perkins AC, Tallack MR, Xu Z, Liu H (2012) Prediction of novel long non-coding RNAs based on RNA-Seq data of mouse Klf1 knockout study. BMC Bioinform 13:331
Sutter JMJ, Kalivas JHJ (1993) Comparison of forward selection, backward elimination, and generalized simulated annealing for variable selection. Microchem J 47:60–66
Thangaiah PR, Shriram R, Vivekanandan K (2009) Adaptive hybrid methods for Feature selection based on Aggregation of Information gain and Clustering methods. Int J Comput Sci Netw Secur 9(2):164–169
Tripathi R, Sharma P, Chakraborty P, Varadwaj PK (2016) Next-generation sequencing revolution through big data analytics. Front Life Sci. doi:10.1080/21553769.2016.1178180
Volders PJ, Helsens K, Wang X, Menten B, Martens L, Gevaert K, Vandesompele J, Mestdaghet P (2013) LNCipedia: a database for annotated human IncRNA transcript sequences and structures. Nucleic Acids Res 41(Database issue):D246–D251
Wager S, Wang S, Liang PC (2013) Dropout training as adaptive regularization. NIPS, pp 1–11
Wain HM, Lush MJ, Ducluzeau F, Khodiyar VK, Povey S (2004) Genew: the human gene nomenclature database. Nucleic Acids Res 32:255–257
Wang Z, Gerstein M, Snyder M (2009) RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet 10(1):57–63
Wapinski O, Chang HY (2011) Long noncoding RNAs and human disease. Trends Cell Biol 21:354–361
Washietl S, Hofacker IL (2007) Identifying structural noncoding RNAs using RNAz. Curr Protoc Bioinformatics. doi:10.1002/0471250953.bi1207s19
Wright MW (2014) A short guide to long non-coding RNA gene nomenclature. Human genomics. BioMed Central Ltd 8(1):7
Xie C, Yuan J, Li H, Li M, Zhao G, Bu D, Zhu W, Wu W, Chen R, Zhao Y (2014) NONCODEv4: exploring the world of long non-coding RNA genes. Nucleic Acids Res 42(Database issue):D98–D103
Yan ZJ, Huo Q, Xu J (2013) A scalable approach to using DNN-derived features in GMM-HMM based acoustic modeling for LVCSR. In: Proceedings of the Annual Conference of the International Speech Communication Association, INTERSPEECH. International Speech and Communication Association, pp 104–108
Yang JH, Li JH, Jiang S, Zhou H, Qu LH (2013) ChIPBasea database for decoding the transcriptional regulation of long non-coding RNA and microRNA genes from ChIP-Seq data. Nucleic Acids Res 41(D):177–187
Zeiler MD (2012) ADADELTA: an adaptive learning rate method. eprint http://arXiv.1212.5701
Zhang Y, Guan DG, Yang JH, Shao P, Zhou H, Qu LH (2010) ncRNAimprint: a comprehensive database of mammalian imprinted noncoding RNAs. RNA 16(10):1889–1901
Zhao J, Ohsumi TK, Kung JT, Ogawa Y, Grau DJ, Sarma K, Song J, Kingston R, Borowsky M, Lee JT (2010) Genome-wide identification of polycomb-associated RNAs by RIP-seq. Mol Cell 40(6):939–953
Zhou M, Wang X, Li J, Hao D, Wang Z, Shi H, Han L, Zhou H, Sun J (2015) Prioritizing candidate disease-related long non-coding RNAs by walking on the heterogeneous lncRNA and disease network. Mol BioSyst 11(3):760–769
Zhu J, Liu S, Ye F, Shen Y, Tie Y, Zhu J, Jin Y, Zheng X, Wu Y, Fu H (2014) The long noncoding RNA expression profile of hepatocellular carcinoma identified by microarray analysis. PLoS One 9(7):e101707. doi:10.1371/journal.pone.0101707
Acknowledgments
We are thankful to Department of Bioinformatics, Indian Institute of Information Technology-Allahabad, India for providing the computational facility to perform the study.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Tripathi, R., Patel, S., Kumari, V. et al. DeepLNC, a long non-coding RNA prediction tool using deep neural network. Netw Model Anal Health Inform Bioinforma 5, 21 (2016). https://doi.org/10.1007/s13721-016-0129-2
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13721-016-0129-2