Abstract
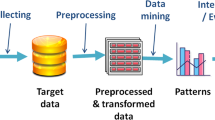
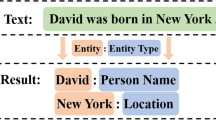
One of the important subtasks of information extraction is named entity recognition (NER). Its aim is to identify and to classify the named entities in the textual data into predetermined categories. There are a large number of supervised learning and deep learning models being developed for the entity recognition task, which performs well in the presence of a labeled training set. The availability of the labeled training set requires the labeling of large unlabeled data, which is both expensive and time taking. Active learning is an iterative approach that provides a way to minimize labeling cost without affecting performance. This approach uses a sampling strategy that selects the appropriate unlabeled data instances, an oracle to label the selected data instances, and a machine learning model (base classifier). In this work, a modified least confidence-based query sampling strategy for the active learning approach for named entity recognition task has been proposed, which considers different numbers of uncertain words present within the sentences to compute the final least confidence score of the sentence for comparison. To evaluate the effectiveness of the proposed approach, the comparison of the performance is made among the active learning approaches with the proposed sampling strategy, random sampling strategy, and two other well-known existing uncertainty query sampling strategies. Real-world scenario for active learning approach is simulated for experiment, and the total amount of labeled data required for training of active learner to reach the stop condition while using different sampling strategies is recorded. The experiment is carried for the development and the test set of the three different biomedical corpora and a Spanish language NER corpus. It is found that with the proposed active learning approach, there is a minimal requirement of labeled data for training to reach the above performance level in comparison with the other approaches. The performance of the proposed approach is found to be slightly better than the existing sampling approach, and the performance of all the approaches is far better than the random sampling approach.




Similar content being viewed by others
References
PMC Repository Information. https://www.ncbi.nlm.nih.gov/pmc/. Accessed 03 Aug 2019.
Benajiba, Y., Rosso, P., Lyhyaoui, A.: Implementation of the ArabiQA question answering system’s components. In: Proceedings of the 2nd Information Communication Technologies International Symposium Workshop on Arabic Natural Language Processing, ICTIS-2007, pp. 3–5. Fez, Morroco (2007).
Abdi, A., Hasan, S., Arshi, M., Shamsuddin, S.M., Idris, N.: A question answering system in hadith using linguistic knowledge. Comput. Speech Lang. (2019). https://doi.org/10.1016/j.csl.2019.101023
Trisedya, B.D., Weikum, G., Qi, J., Zhang, R.: Neural relation extraction for knowledge base enrichment. In: Proceedings of the 57th Annual Meeting of the Association for Computational Linguistics, pp. 229–240. Association for Computational Linguistics, Florence, Italy (2019). https://doi.org/10.18653/v1/P19-1023.
Khalifa, M., Shaalan, K.: Character convolutions for Arabic named entity recognition with long short-term memory networks. Comput. Speech Lang. 58, 335–346 (2019). https://doi.org/10.1016/j.csl.2019.05.003
Aguilar, G., Maharjan, S., López-Monroy, A.P., Solorio, T.: A multi-task approach for named entity recognition in social media data. CoRR. abs/1906.0 (2019).
Yeniterzi, R., Tür, G., Oflazer, K.: Turkish named-entity recognition. In: Oflazer, K., Saraçlar, M. (eds.) Turkish Natural Language Processing, pp. 115–132. Springer, Cham (2018). https://doi.org/10.1007/978-3-319-90165-7_6.
Li, J., Sun, A., Han, J., Li, C.: A survey on deep learning for named entity recognition. CoRR. abs/1812.0 (2018).
Nadeau, D., Sekine, S.: A survey of named entity recognition and classification. Lingvisticae Investig. 30, 3–26 (2007). https://doi.org/10.1075/li.30.1.03nad
Krishnan, V., Manning, C.D.: An effective two-stage model for exploiting non-local dependencies in named entity recognition. In: Proceedings of the 21st International Conference on Computational Linguistics and 44th Annual Meeting of the Association for Computational Linguistics, pp. 1121–1128. Association for Computational Linguistics, Sydney, Australia (2006). https://doi.org/10.3115/1220175.1220316.
Sang, K.T.E.F., De Meulder, F.: Introduction to the CoNLL-2003 shared task: language-independent named entity recognition. In: Proceedings of the Seventh Conference on Natural Language Learning at HLT-NAACL, pp. 142–147. Association for Computational Linguistics (2003).
Kazama, J., Makino, T., Ohta, Y., Tsujii, J.: Tuning support vector machines for biomedical named entity recognition. In: Proceedings of the ACL-02 Workshop on Natural Language Processing in the Biomedical Domain, vol 3, pp. 1–8. Association for Computational Linguistics, Stroudsburg (2002). https://doi.org/10.3115/1118149.1118150.
Shen, Y., Yun, H., Lipton, Z.C., Kronrod, Y., Anandkumar, A.: Deep active learning for named entity recognition. CoRR. abs/1707.0 (2017).
Zhao, Z., Yang, Z., Luo, L., Wang, L., Zhang, Y., Lin, H., Wang, J.: Disease named entity recognition from biomedical literature using a novel convolutional neural network. BMC Med. Genomics. 10, 73 (2017). https://doi.org/10.1186/s12920-017-0316-8
Campos, D., Matos, S., Oliveira, J.L.: Biomedical named entity recognition: a survey of machine-learning tools. In: Sakurai, S. (ed.) Theory and Applications for Advanced Text Mining. IntechOpen, Rijeka (2012). https://doi.org/10.5772/51066.
Chang, K.H.: Explaining active learning queries (2017).
Chen, Y., Lasko, T.A., Mei, Q., Denny, J.C., Xu, H.: A study of active learning methods for named entity recognition in clinical text. J. Biomed. Inform. 58, 11–18 (2015). https://doi.org/10.1016/j.jbi.2015.09.010.
Ekbal, A., Saha, S., Sikdar, U.K.: On active annotation for named entity recognition. Int. J. Mach. Learn. Cybern. 7, 623–640 (2016). https://doi.org/10.1007/s13042-014-0275-8
Liu, M., Tu, Z., Wang, Z., Xu, X.: LTP: a new active learning strategy for bert-crf based named entity recognition (2020).
Huang, H., Wang, H., Jin, D.: A low-cost named entity recognition research based on active learning. Sci. Program. 2018, 10 (2018). https://doi.org/10.1155/2018/1890683
Skeppstedt, M., Paradis, C., Kerren, A.: PAL: a tool for pre-annotation and active learning. J. Lang. Technol. Comput. Linguist. 31, 91–110 (2017)
Klie, J.-C.: INCEpTION: interactive machine-assisted annotation. In: Proceedings of the First Biennial Conference on Design of Experimental Search and Information Retrieval Systems. p. 105 (2018).
Klie, J.-C., Bugert, M., Boullosa, B., de Castilho, R.E., Gurevych, I.: The INCEpTION platform: machine-assisted and knowledge-oriented interactive annotation. In: Proceedings of the 27th International Conference on Computational Linguistics: System Demonstrations, pp. 5–9. Association for Computational Linguistics (2018).
Kholghi, M., Sitbon, L., Zuccon, G., Nguyen, A.: Active learning reduces annotation time for clinical concept extraction. Int. J. Med. Inform. 106, 25–31 (2017). https://doi.org/10.1016/j.ijmedinf.2017.08.001
Van Tran, C., Nguyen, T.T., Hoang, D.T., Hwang, D., Nguyen, N.T.: Active learning-based approach for named entity recognition on short text streams. In: Zgrzywa, A., Choroś, K., Siemiński, A. (eds.) Multimedia and Network Information Systems, pp. 321–330. Springer, Cham (2017)
Tran, V.C., Hoang, D.T., Nguyen, N.T., Hwang, D.: A hybrid method for named entity recognition on tweet streams. In: Nguyen, N.T., Tojo, S., Nguyen, L.M., Trawiński, B. (eds.) Intelligent Information and Database Systems, pp. 258–268. Springer, Cham (2017)
Project, G.: BioNLP/JNLPBA Shared Task 2004. http://www.geniaproject.org/shared-tasks/bionlp-jnlpba-shared-task-2004.
Collier, N., Kim, J.-D.: Introduction to the bio-entity recognition task at JNLPBA. In: Proceedings of the International Joint Workshop on Natural Language Processing in Biomedicine and its Applications ({NLPBA}/{B}io{NLP}), pp. 73–78. COLING, Geneva (2004).
Kim, J.-D., Ohta, T., Tateisi, Y., Tsujii, J.: GENIA corpus—a semantically annotated corpus for bio-textmining. Bioinformatics 19, i180–i182 (2003)
Crichton, G., Pyysalo, S., Chiu, B., Korhonen, A.: A neural network multi-task learning approach to biomedical named entity recognition. BMC Bioinform. 18, 368 (2017). https://doi.org/10.1186/s12859-017-1776-8
Doğan, R.I., Leaman, R., Lu, Z.: NCBI disease corpus: a resource for disease name recognition and concept normalization. J. Biomed. Inform. 47, 1–10 (2014). https://doi.org/10.1016/j.jbi.2013.12.006
Li, J., Sun, Y., Johnson, R.J., Sciaky, D., Wei, C.-H., Leaman, R., Davis, A.P., Mattingly, C.J., Wiegers, T.C., Lu, Z.: BioCreative V CDR task corpus: a resource for chemical disease relation extraction. Database. 2016, (2016). https://doi.org/10.1093/database/baw068.
Bhasuran, B., Murugesan, G., Abdulkadhar, S., Natarajan, J.: Stacked ensemble combined with fuzzy matching for biomedical named entity recognition of diseases. J. Biomed. Inform. 64, 1–9 (2016). https://doi.org/10.1016/j.jbi.2016.09.009
Tjong Kim Sang, E.F.: Introduction to the CoNLL-2002 shared task: language-independent named entity recognition. In: Proceedings of the 6th Conference on Natural Language Learning, vol 20, pp. 1–4. Association for Computational Linguistics, Stroudsburg (2002). https://doi.org/10.3115/1118853.1118877.
Korobov, M.: sklearn-crfsuite docs. https://sklearn-crfsuite.readthedocs.io/en/latest/tutorial.html. Accessed 04 Nov 2019.
Classification: True vs. false and positive vs. negative. https://developers.google.com/machine-learning/crash-course/classification/true-false-positive-negative.
Settles, B.: From theories to queries: active learning in practice. Active Learning and Experimental Design workshop In conjunction with AISTATS 2010, 1–18 (2011)
Settles, B.: Active learning. Synth. Lect. Artif. Intell. Mach. Learn. 6, 1–114 (2012). https://doi.org/10.2200/S00429ED1V01Y201207AIM018
Lewis, D.D., Catlett, J.: Heterogeneous uncertainty sampling for supervised learning. In: Cohen, W.W., Hirsh, H. (eds.) Machine Learning Proceedings 1994, pp. 148–156. Morgan Kaufmann, San Francisco (1994). https://doi.org/10.1016/B978-1-55860-335-6.50026-X.
Culotta, A., McCallum, A.: Reducing Labeling effort for structured prediction tasks. In: Proceedings of the 20th National Conference on Artificial Intelligence, vol 2, pp. 746–751. AAAI Press, Palo Alto (2005).
Settles, B., Craven, M.: An analysis of active learning strategies for sequence labeling tasks. In: Proceedings of the Conference on Empirical Methods in Natural Language Processing. pp. 1070–1079. Association for Computational Linguistics, Stroudsburg (2008).
Lin, Y., Sun, C., Xiaolong, W., Xuan, W.: Combining Self Learning and Active Learning for Chinese Named Entity Recognition. J. Softw. 5, (2010). https://doi.org/10.4304/jsw.5.5.530-537.
Laws, F., Schätze, H.: Stopping criteria for active learning of named entity recognition. In: Proceedings of the 22Nd International Conference on Computational Linguistics, vol 1, pp. 465–472. Association for Computational Linguistics, Stroudsburg (2008).
Vlachos, A.: A stopping criterion for active learning. Comput. Speech Lang. 22, 295–312 (2008). https://doi.org/10.1016/j.csl.2007.12.001
Confidence-based active learning: Mingkun Li, Sethi, I.K. IEEE Trans. Pattern Anal. Mach. Intell. 28, 1251–1261 (2006). https://doi.org/10.1109/TPAMI.2006.156
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Agrawal, A., Tripathi, S. & Vardhan, M. Active learning approach using a modified least confidence sampling strategy for named entity recognition. Prog Artif Intell 10, 113–128 (2021). https://doi.org/10.1007/s13748-021-00230-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13748-021-00230-w