Abstract
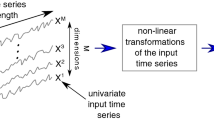
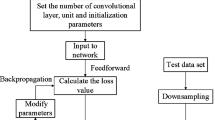
The application of various sensors in hospitals has enabled the widespread utilization of multivariate time series signals for chronic disease diagnosis in the data-driven world. The key challenge is how to model the complex temporal (linear and nonlinear) correlations among multiple longitudinal variables. Due to scarcity of labels in practice, unsupervised learning methods have already become indispensable. However, state-of-the-art approaches mainly focus on the extraction of linear correlation-induced feature connectivity network, e.g., Pearson correlation, partial correlation, etc. To this end, for chronic disease (e.g., Parkinson disease) diagnosis, an unsupervised representation learning method is first designed to obtain informative correlation-aware signals from multivariate time series data. At the core is a contrastive learning framework with a graph neural network (GNN) encoder to capture the inter-correlation and intra-correlation of multiple longitudinal variables. Then, the previously learned representations are sent to a simple fully connected neural network, which can be trained using fewer labels compared with end-to-end complex supervised learning models. Further, to assist the decision-making process in the high-stake chronic disease detection task, model uncertainty quantification is enabled according to evidential theory. The experimental results on two public Parkinson’s disease data sets show the expressiveness of the learned embeddings, and the final lightweight classifier achieves the best performance.





Similar content being viewed by others
References
Killin, L., Starr, J., Shiue, I., Russ, T.: Environmental risk factors for dementia: a systematic review. BMC Geriatr. 1, 175 (2016)
Thomson, R.: Disease briefing: Parkinson’s disease. J. Int. Pharm. Res. 3, 338–345 (2015)
Futoma, J., Sendak, M., Cameron, B.: Predicting disease progression with a model for multivariate longitudinal clinical data. Proc. Mach. Learn. Res. 56, 42–54 (2016)
Shen, X., Finn, E.S., Scheinost. D., Rosenberg, M.D., Chun, M.M., Pa-pademetris, X., Constable, R.T.: Using connectome-based predictive modeling to predict individual behavior from brain connectivity, nature protocols, vol. 12(3), p. 506 (2017).
Yu, R., Qiao, L., Chen, M., Lee, S.W., Fei, X., Shen, D.: Weighted graph regularized sparse brain network construction for mci identification. Pattern Recogn. 90, 220–231 (2019)
Huang, H., Liu, X., Jin, Y., Lee, S.W., Wee, C.Y., Shen, D.: Enhancing the representation of functional connectivity networks by fusing multi-view information for autism spectrum disorder diagnosis. Hum. Brain Mapp. 40(3), 833–854 (2019)
Qu, Y., Liu, C.R., Zhang, K., Xiao K., Jin, B., Xiong, H.: Diagnostic sparse connectivity networks with regularization template. IEEE Trans. Knowl. Data Eng. (TKDE) (2021).
Jin, B., Cheng, K., Qu, Y., Zhang, L., Xiao, K.L., Lu, X.J., Wei, X.P.: Fast sparse connectivity network adaption via meta-learning. In: ICDM’20. pp.232–241(2020).
Wu, Z., Pan, S., Long, G.: Connecting the dots: multivariate time series forecasting with graph neural networks. In: ACM’20 (2020).
Zhang, X., Gao, Y., Lin, J., Lu, C.T.: Tap-net: multivariate time series classification with attentional prototypical network. In: AAAI’20, pp. 6845–6852 (2020).
Shokoohi, M., Wang, J., Keogh, E.: On the non-trivial generalization of dynamic time warping to the multi-dimensional case. In: Proceedings of the 2015 SIAM International Conference on Data Mining, pp. 289–297 (2015).
Karim, F., Majumdar, S., Darabi, H., Harford, S.: Multivariate LSTM-FCNs for time series classification. Neural Netw. 116, 237–245 (2019)
Jin, B., Qu, Y., Zhang, L., Gao, Z.: Diagnosing Parkinson disease through facial expression recognition: video analysis. J. Med. Internet Res. (2020). https://doi.org/10.2196/18697
Bandini, A., Orlandi, S., Escalante, H.J., Giovannelli. F., Cincotta, M., Reyes C.A.: Analysis of facial expressions in Parkinson's disease through video-based automatic methods. J. Neurosci. Methods (2017). https://doi.org/10.1016/j.jneumeth.2017.02.006
Rajnoha, M., Mekyska, J., Burget, R., Eliasova, I., Kostalova, M., Rektorova, I.: Towards identification of hypomimia in Parkinson's disease based on face recognition methods. In: 10th International Congress on Ultra Modern Telecommunications and Control Systems and Workshops (ICUMT) (2018). pp. 1–4. https://doi.org/10.1109/ICUMT.2018.8631249
Dustin. T., Mike, D., Mark, V.D., Danijar, H.: Bayesian layers: a module for neural network uncertainty. In: Advances in Neural Information Processing Systems, pp. 14660–14672 (2019)
Murat, S., Lance, K., Melih, K.: Evidential deep learning to quantify classification uncertainty. In: Advances in Neural Information Processing Systems 31, pp. 3179–3189 (2018).
Goh, K.I., Cusick, M.E., Valle, D., Childs, B., Vidal, M., Barabási, A.: The human disease network. In: Proceedings of the National Academy of Sciences(PNAS), vol. 104, no. 21, pp. 8685–8690 (2007)
Wang, K., Liang, M., Wang, L., Tian, L., Zhang, X., Li, K., Jiang, T.: Altered functional connectivity in early alzheimer’s disease: a resting-state fmri study. Hum. Brain Mapp. 28(10), 967–978 (2007)
Banerjee, O., Ghaoui, L.E., Aspremont, A., Natsoulis, G.: Convex optimization techniques for fitting sparse Gaussian graphical models. In: Proceedings of the 23rd International Conference on Machine Learning (ICML). ACM, pp. 89–96 (2006).
Lee, H., Lee, D.S., Kang, H., Kim, B., Chung, M.K.: Sparse brain network recovery under compressed sensing. IEEE Trans. Med. Imaging 30(5), 1154–1165 (2011)
Candès, E.J., Wakin, M.B.: An introduction to compressive sampling, In: IEEE Signal Processing Magazine, vol. 25, no. 2, pp. 21–30 (2008)
Lei, Q., Yi, J., Vaculin, R.: Similarity preserving representation learning for time series clustering. In: International Joint Conferences on Artificial Intelligence Organization, pp. 2845–2851 (2017)
Malhotra, P., Tv, V., Vig, L.: TimeNet: pre-trained deep recurrent neural network for time series classification. In: 25th European Symposium on Artificial Neural Networks, Computational Intelligence and Machine Learning (2017)
Franceschi, J.Y.: Unsupervised Scalable representation learning for multivariate time series. In: Thirty Third Conference on Neural Information Processing Systems, vol. 32, pp. 4650–4661 (2019)
Wu, Z., Pan, S., Chen, F., Long, G., Zhang, C., Yu, P.S.: A comprehensive survey on graph neural networks. In: IEEE Transactions on Neural Networks and Learning Systems (2019).
Scarselli, F., Gori, M., Tsoi, C.A., Hagenbuchner, M., Monfardini, G.: The graph neural network model. IEEE Trans. Neural Netw. 20(1), 61 (2009)
Wu, Z., Pan, S., Long, G., Jiang, J., Chang, X., Zhang, C.: Connecting the dots: multivariate time series forecasting with graph neural networks. In: The 26th ACM SIGKDD Conference on Knowledge Discovery and Data Mining. KDD, pp. 753–763 (2020)
Spadondesouza, G., Hong, S., Brandoli, B.: Pay attention to evolution: time series forecasting with deep graph-evolution learning. IEEE Trans. Pattern Anal. Mach. Intell. (2021)
Du, J., Zhang, S.H., Wu, G.H., Moura, J., Kar, S.: Topology adaptive graph convolutional networks. In: International Conference on Learning Representations. ICLR (2018)
Yikuan, L., Shishir, R., Abdelaali, H., Rema, R., Zhu, Y.J., Dexter C., Gholamreza S., Thomas L., Kazem, R.: Deep Bayesian Gaussian Processes for Uncertainty Estimation in Electronic Health Records (2020). arXiv preprint arXiv:2003.10170
Zhang, C., Bütepage, J., Kjellström, H., Mandt, S.: Advances in variational inference. IEEE Trans. Pattern Anal. Mach. Intell. 41, 2008–2026 (2019)
Sakar, B.E., Isenkul, M.E., Sakar, C.O., Sertbas, A., Gürgen, F.S., Delil, S., Apaydin, H., Kursun, O.: Collection and analysis of a parkinson speech dataset with multiple types of sound recordings. IEEE J. Biomed. Health Inform. 17, 828–834 (2013)
Dempster, A.P.: A generalization of Bayesian inference. In: Classic Works of the Dempster–Shafer Theory of Belief Functions. Springer, pp. 73–104 (2008).
Josang, A.: Subjective Logic: A Formalism for Reasoning Under Uncertainty. Springer, Berlin (2016)
Kotz, S., Balakrishnan, N., Johnson, N.L.: Continuous Multivariate Distributions. Wiley, New York (2000)
Maaten, L.V., Hinton, G.: Visualizing data using t-SNE. J. Mach. Learn. Res. 9, 2579–2605 (2008)
Goodfellow, I. J., Shlens, J., Szegedy, C.: Explaining and harnessing adversarial examples. Comput. Sci. (2014)
Szegedy, C., Liu, W., Jia, Y. Q.: Going deeper with convolutions. In: Conference on Computer Vision and Pattern Recognition. CVPR (2015)
Sensoy, M., Kaplan, L., Kandemir, M.: Evidential deep learning to quantify classification uncertainty, In: Advances in Neural Information Processing Systems (2018)
Wang, G.F., Cao, L.B., Zhao, H.K., Liu, Q., Chen, E.H.: Coupling macro-sector-micro financial indicators for learning stock representations with less uncertainty. In The 35th AAAI Conference on Artificial Intelligence. pp. 4418–4426 (2021)
Sakar, B.E., Isenkul, M.E., Sakar, C.O., Sertbas, O., et al.: Collection and analysis of a Parkinson speech dataset with multiple types of sound recordings. IEEE J. Biomed. Health Inform. 17(4), 828–834 (2013)
Little, M.A.P.E., McSharry, E.J., Spielman, H.J., Ramig, L.O.: Suitability of dysphonia measurements for telemonitoring of Parkinson’s disease. IEEE Trans. Biomed. Eng. 56(4), 1010–1022 (2009)
Sakar, C.O., Kursun, O.: Telediagnosis of Parkinson’s disease using measurements of dysphonia. J. Med. Syst. 34(4), 591–599 (2010)
Funding
This research was funded by National Natural Science Foundation of China, Grant Number 61772110, and Key Program of Liaoning Traditional Chinese Medicine Administration, Grant Number LNZYXZK201910.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Zhang, X., Wang, Y., Zhang, L. et al. Exploring unsupervised multivariate time series representation learning for chronic disease diagnosis. Int J Data Sci Anal 15, 173–186 (2023). https://doi.org/10.1007/s41060-021-00290-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s41060-021-00290-0