Abstract
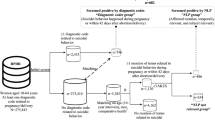
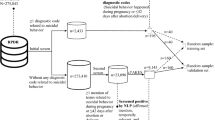
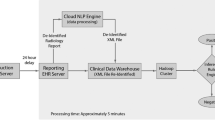
Accurate and complete identification of spontaneous abortion (SAB) is critical important for conducting SAB-related studies. We developed and validated a natural language processing (NLP) algorithm to identify SAB from free-text clinical notes. Potential SAB cases were identified among subjects after influenza vaccinations during influenza season of 2012–2015 or quadrivalent human papillomavirus (4vHPV) vaccinations in 2012–2013 through diagnosis codes. The SAB and the matched non-SAB subjects after influenza vaccinations were used to develop the NLP algorithm. Chart-reviewed and adjudicated SAB cases after 4vHPV vaccinations were used to validate the algorithm performance. The developed algorithm was then applied to documented pregnancy episodes in the electronic medical record (EMR) system in 2011–2014. The NLP results were compared against the cases identified by diagnosis codes. The NLP algorithm successfully identified 289 of the 310 confirmed SAB cases in the validation dataset and achieved a sensitivity of 93.2% (95% confidence interval [CI] 90.4–96.0%), positive predictive value of 96.0% (95% CI 93.8–98.2%)specificity of 58.6% (95% CI 40.7–76.6%) and negative predictive value of 44.7% (95% CI 28.9–60.6%) when it was applied to the potential SAB cases identified by diagnosis codes in the 4vHPV validation dataset. When the NLP process was applied to the 195,395 documented pregnancies between 2011 and 2014, the NLP algorithm identified a total of 20,709 potential SAB cases (10.6% of pregnancies). Of those potential cases, 9856 (47.6%) were not identified by diagnosis codes. These data suggest that the NLP algorithm can be used to identify SAB from EMR effectively. The developed algorithms could be potentially augmented with EMR structured data to improve the accuracy.

Similar content being viewed by others
References
Loss EP. Practice bulletin no. 150, American college of obstetricians and gynecologists. Obstet Gynecol. 2015;125(9):1258–67.
Wilcox AJ, Weinberg CR, O’Connor JF, Baird DD, Schlatterer JP, Canfield RE, et al. Incidence of early loss of pregnancy. N Engl J Med. 1988;319:189–94.
Brigham SA, Conlon C, Farquharson RG. A longitudinal study of pregnancy outcome following idiopathic recurrent miscarriage. Hum Reprod. 1999;14:2868–71.
Griebel CP, Halvorsen J, Golemon TB, Day AA. Management of spontaneous abortion. Am Fam Physician. 2005;72:1243–50.
Ellett K, Buxton EJ, Luesley DM. The effect of ethnic origin on the rate of spontaneous late mid-trimester abortion. Ethn Dis. 1992;2(1):84–6.
Ammon Avalos L, Galindo C, Li D-K. A systematic review to calculate background miscarriage rates using life table analysis. Birth Defects Res A Clin Mol Teratol. 2012;94(6):417–23. https://doi.org/10.1002/bdra.23014.
Mercer B, Milluzzi C, Collin M. Periviable birth at 20 to 26 weeks of gestation: proximate causes, previous obstetric history and recurrence risk. Am J Obstet Gynecol. 2005;193(3 pt 2):1175–80.
Stephenson MD, Awartani KA, Robinson WP. Cytogenetic analysis of miscarriages from couples with recurrent miscarriage: a case-control study. Hum Reprod. 2002;17:446–51.
Risch HA, Weiss NS, Clarke EA, Miller AB. Risk factors for spontaneous abortion and its recurrence. Am J Epidemiol. 1988;128(2):420–30.
Floyd RL, Jack BW, Cefalo R, Atrash H, Mahoney J, Herron A, et al. The clinical content of preconception care: alcohol, tobacco, and illicit drug exposures. Am J Obstet Gynecol. 2008;199:S333–9.
Lashen H, Fear K, Sturdee DW. Obesity is associated with increased risk of first trimester and recurrent miscarriage: matched case–control study. Hum Reprod. 2004;19(7):1644–6.
Mills JL, Simpson JL, Driscoll SG, Jovanovic-Peterson L, Van Allen M, Aarons JH, et al. Incidence of spontaneous abortion among normal women and insulin-dependent diabetic women whose pregnancies were identified within 21 days of conception. N Engl J Med. 1988;319(25):1617–23.
Harper SA, Fukuda K, Uyeki TM, Cox NJ, Bridges CB. Prevention and control of influenza: recommendations of the advisory committee on immunization practices (ACIP). MMWR Recomm Rep. 2004;53:1–40.
ACOG Committee on Obstetric Practice. Opinion No 732: influenza vaccination during pregnancy. Obstet Gynecol. 2018;131:e109–14.
Lieu TA, Nguyen MD, Ball R, Martin DB. Health outcomes of interest for eval-uation in the Post-Licensure Rapid Immunization Safety Monitoring Program. Vaccine. 2012;30:2824–30.
Baggs J, Gee J, Lewis E, Fowler G, Benson P, Lieu T, et al. The Vaccine Safety Datalink: a model for monitoring immunization safety. Pediatrics. 2011;127(Suppl 1):S45-53.
Irving SA, Kieke BA, Donahue JG, Mascola MA, Baggs J, DeStefano F, et al. Vaccine Safety Datalink. Trivalent inactivated influenza vaccine and spontaneous abortion. Obstet Gynecol. 2013;121(1):159–65.
Donahue JG, Kieke BA, King JP, DeStefano F, Mascola MA, Irving SA, et al. Association of spontaneous abortion with receipt of inactivated influenza vaccine containing H1N1pdm09 in 2010–11 and 2011–12. Vaccine. 2017;35:5314–22.
Donahue JG, Kieke BA, King JP, Mascola MA, Shimabukuro TT, DeStefano F, et al. Inactivated influenza vaccine and spontaneous abortion in the Vaccine Safety Datalink in 2012–13, 2013–14, and 2014–15. Vaccine. 2019;37(44):6673–7668. https://doi.org/10.1016/j.vaccine.2019.09.035.
Kharbanda EO, Vazquez-Benitez G, Lipkind HS, Sheth SS, Zhu J, Naleway AL, et al. Risk of spontaneous abortion after inadvertent human papillomavirus vaccination in pregnancy. Obstet Gynecol. 2018;132:35–44.
Likis FE, Sathe NA, Carnahan R, McPheeters ML. A systematic review of validated methods to capture stillbirth and spontaneous abortion using administrative or claims data. Vaccine. 2013;31:K74–82.
Sneider K, Langhoff-Roos J, Sundtoft IB, Christiansen OB. Validation of second trimester miscarriages and spontaneous deliveries. Clin Epidemiol. 2017;9:367–8.
Lohse SR, Farkas DK, Lohse N, Skouby SO, Nielsen FE, Lash TL, et al. Validation of spontaneous abortion diagnoses in the Danish National Registry of Patients. Clin Epidemiol. 2010;2:247–50.
Araneta MR, Kamens DR, Zau AC, Gastanaga VM, Schlangen KM, Hiliopoulos KM, et al. Conception and pregnancy during the Persian Gulf War: the risk to women veterans. Ann Epidemiol. 2004;14(2):109–16.
Mikolajczyk RT, Kraut AA, Garbe E. Evaluation of pregnancy outcome records in the German Pharmacoepidemiological Research Database (GePaRD). Pharmacoepidemiol Drug Saf. 2013;22:873–80.
Fridman C, Alderson PO, Austin J, Cimino JJ, Johnson SB. A general natural language text processor for clinical radiology. J Am Med Inform Assoc. 1994;1:161–74.
Jensen PB, Jensen LJ, Brunak S. Mining electronic health records: towards better research applications and clinical care. Nat Rev Genet. 2012;13:395–405.
Savova GK, Masanz JJ, Ogren PV, Zheng J, Sohn S, Kipper-Schuler KC, et al. Mayo clinical Text Analysis and Knowledge Extraction System (cTAKES): architecture, component evaluation and applications. J Am Med Inform Assoc. 2010;17:507–13.
Crowley RS, Castine M, Mitchell K, Chavan G, McSherry T, Feldman M. CaTIES: A grid based system for coding and retrieval of surgical pathology reports and tissue specimens in support of translational research. J Am Med Inform Assoc. 2010;17:253–64.
Chapman B, Chapman WW, Dayton G, Mowery D. Python implementation of the ConText Algorithm. Available from: https://github.com/chapmanbe/pyConTextNLP/ [Last accessed on 2019 Apr 1]
Xu H. Clinical language annotation, modeling, and processing toolkit. Available from: http://clamp.uth.edu/. [Last accessed on 2019 Apr 1].
Wang X, Hripcsak G, Markatou M, Friedman C. Active computerized pharmacovigilance using natural language processing, statistics, and electronic health records: a feasibility study. J Am Med Inform Assoc. 2009;16:328–37.
Botsis T, Buttolph T, Nguyen MD, Winiecki S, Woo EJ, Ball R. Vaccine adverse event text mining system for extracting features from vaccine safety reports. J Am Med Inform Assoc. 2012;19:1011–8.
Zheng C, Yu W, Xie F, Chen W, Mercado C, Sy LS, et al. The use of natural language processing to identify Tdap-related local reactions at five health care systems in the Vaccine Safety Datalink. IJMI. 2019;127:27–34.
Yu W, Zheng C, Xie F, Chen W, Mercado C, Sy LS, et al. the use of natural language processing to identify vaccine-related anaphylaxis at five health care systems in the vaccine safety datalink. Accepted by the PDS journal
Koebnick C, Langer-Gould AM, Gould MK, Chao CR, Lyer RL, Smith N, et al. Sociodemographic characteristics of members of a large, integrated health care system: comparison with US Census Bureau data. Permanente J. 2012;16:37–41.
Naleway AL, Gold R, Kurosky S, Reidlinger K, Henninger ML, Nordin JD, et al. Identifiying pregnancy episodes, outcomes, and mother-infant pairs in the Vaccine Safety Datalink. Vaccine. 2013;31(27):2898–903.
Griffon N, Chebil W, Rollin L, Kerdelhue G, Thirion B, Gehanno JF, et al. Performance evaluation of unified medical language system®’s synonyms expansion to query PubMed. BMC Med Inform Decis Mak. 2012;12:12.
Loper E, Bird S. NLTK: the Natural Language Toolkit, Proceedings of the ACL-02 Workshop on Effective tools and methodologies for teaching natural language processing and computational linguistics, pp. 63–70, July 07–07, 2002, Philadelphia, Pennsylvania
Xie F, Im T, Getahun D. A computerized algorithm to capture patient’s past preeclampsia and eclampsia history from prenatal clinical notes. Health Inform J. 2019;25(4):1299–313.
Altman DG, Bland JM. Diagnostic tests 2: predictive values. BMJ. 1994;309:102.
Getahun D, Rhoads GG, Fassett MJ, Chen W, Strauss JA, Demissie K, et al. Accuracy of reporting maternal and infant perinatal service system coding and clinical utilization coding. J Med Stat Inform. 2013;1(1):1–3.
Mikolov T, Chen K, Corrado G, Dean J. Efficient estimation of word representations in vector space. In: 1st International Conference on Learning Representations (ICLR) 2013—Workshop Track Proceedings
Pennington J, Socher R, Manning CD. Glove: global vectors for word representation. In: Proceedings of the 2014 Conference on Empirical Methods in Natural Language Processing (EMNLP), 2014;31(6):1532–1543. https://doi.org/10.3115/v1/D14-1162.
Bojanowski P, Grave E, Joulin A, Mikolov T. Enriching word vectors with subword information. Trans Assoc Comput Linguistics. 2017;5:135–46. https://doi.org/10.1162/tacl_a_00051.
Sheikhalishahi S, Miotto R, Dudley JT, Lavelli A, Finaldi F, Osman V. Natural language processing of clinical notes on chronic diseases: systematic review. JMIR Med Inform. 2019;7(2): e12239.
Shivade C, Raghavan P, Fosler-Lussier E, Embi PJ, Elhadad N, Johnson SB, et al. A review of approaches to identifying patient phenotype cohorts using electronic health records. J Am Med Inform Assoc. 2014;21(2):221–30.
Castro SM, Tseytlin E, Medvedeva O, Mitchell K, Visweswaran S, Bekhuis T, et al. Automated annotation and classification of BI-RADS assessment from radiology reports. J Biomed Inform. 2017;69:177–87.
The Shared Tasks for Challenges in NLP. Available from: https://www.i2b2.org/NLP/DataSets/Main.php. [Last accessed on 2020 May 20].
The MIMIC-III Critical Care Database. Available from: https://mimic.physionet.org/about/mimic/. [Last accessed on 2020 May 20].
Acknowledgements
Vaccine Safety Datalink (VSD) Project; Kaiser Permanente Southern California: Bernadine Dizon, Bianca Cheung, Claire Park, Cheryl Carlson, Denison Ryan, Farihah Chowdhury, Gina Lee, Joy Gelfond, Kerresa Morrissette, Karen Schenk, Lindsay Joe Lyons, Lee Tillman, Nancy Canul-Jauriga, Nancy Cannizzaro, Radha Bathala, and Sunhea Kim; Yale School of Medicine: Dr. Heather Lipkind and Dr. Sangini Sheth; Marshfield Clinic Research Institute: Dr. Jim Donahue, Dr. Maria Mascola, Jennifer King, Kayla Hanson, and Oluwatosin Olaiya.
Funding
This study was partly supported by Kaiser Permanente Direct Community Benefit Funds. The opinions expressed are solely the responsibility of the authors and do not necessarily reflect the official views of the Kaiser Permanente Community Benefit Funds. The chart review and adjudication work used from previously published VSD studies were funded through the Vaccine Safety Datalink from the Centers for Disease Control and Prevention. The findings and conclusions in this article are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention.
Author information
Authors and Affiliations
Contributions
FX, CM and WC conceived and led the design of the study and drafted the manuscript. FX extracted the data and conducted all of the analysis. CM and SSK conducted all necessary chart reviews of the study datasets. All authors participated in the design of the study, interpreted the results, critically reviewed and revised the manuscript and have given final approval to the manuscript.
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Xie, F., Mercado, C., Kim, S.S. et al. Identifying Spontaneous Abortion from Clinical Notes within a Large Integrated Healthcare System. SN COMPUT. SCI. 3, 268 (2022). https://doi.org/10.1007/s42979-022-01175-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s42979-022-01175-0