Abstract
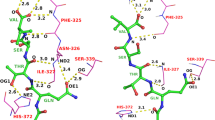
An increasing number of docking/scoring programs are available that use different sampling and scoring algorithms. A reliable scoring function is the crucial element of such approaches. Comparative studies are needed to evaluate their current capabilities. DOCK4 with force field and PMF scoring as well as FlexX were used to evaluate the predictive power of these docking/scoring approaches to identify the correct binding mode of 61 MMP-3 inhibitors in a crystal structure of stromelysin and also to rank them according to their different binding affinities. It was found that DOCK4/PMF scoring performs significantly better than FlexX and DOCK4/FF in both ranking ligands and predicting their binding modes. Most notably, DOCK4/PMF was the only scoring/docking approach that found a significant correlation between binding affinity and predicted score of the docked inhibitors. However, comparing only those cases where the correct binding mode was identified (scoring highest among sampled poses), FlexX showed the best `fine tuning' (lowest rmsd) in predicted binding modes. The results suggest that not so much the sampling procedure but rather the scoring function is the crucial element of a docking program.
Similar content being viewed by others
References
Greer, J., Erickson, J.W., Baldwin, J.J. and Varney, M.D., J. Med. Chem., 37 (1994) 1035.
Lutz, M.W., Menius, J.A., Choi, T.D., Laskody, R.G., Domanico, P.L., Goetz, A.S. and Saussy, D.L., Drug Discov. Today, 1 (1996) 277.
Dixon, J.S., Proteins, Suppl., 1 (1997) 198.
Kuntz, I.D., Blaney, J.M., Oatley, S.J. and Langridge, R.L., J. Mol. Biol., 161 (1982) 269.
Kuntz, I.D., Science, 257 (1992) 1078.
Ewing, T. and Kuntz, I.D., J. Comput. Chem., 18 (1997) 1175.
Rarey, M., Kramer, B., Lengauer, T. and Klebe, G., J. Mol. Biol., 261 (1996) 470.
Welch, W., Ruppert, J. and Jain, A.N., Chem. Biol., 3 (1996) 449.
Jones, G., Willett, P., Glen, R.C. and Leach, A.R., J. Mol. Biol., 267 (1997) 727.
Goodsell, D.S. and Olson, A.J., Proteins, 8 (1990) 195.
Miller, M.D., Kearsley, S.K., Underwood, D.J. and Sheridan, R.P., J. Comput-Aided Mol. Design, 8 (1994) 153.
Dixon, J.S. and Blaney, J.M., In Martin, Y.C. and Willett, P. (Eds.) Designing Bioactive Molecules. Three-dimensional Techniques and Applications, American Chemical Society, Washington, DC, 1998, p. 175.
Böhm, H.-J., J. Comput.-Aided Mol. Design, 8 (1994) 243.
Aqvist, J., Medina, C. and Samuelsson, J.E., Protein Eng., 7 (1994) 386.
Head, R.D., Smythe, M.L., Oprea, T.L., Waller, C.L., Green, S.M. and Marshall, G.M., J. Am. Chem. Soc., 118 (1996) 3959.
Jain, A.N., J. Comput.-Aided Mol. Design, 10 (1996) 427.
Kollman, P., Chem. Rev., 7 (1993) 2395.
Wallqvist, A., Jernigan, R.L. and Covell, D.G., Protein Sci., 4 (1995) 1881.
Williams, D.H., Cox, J.P.L., Doig, A.J., Gardner, M., Gerhard, U., Kaye, P.T., Lai, A.R., Nicholls, I.A., Salter, C.J. and Mitchell, R.C., J. Am. Chem. Soc., 113 (1991) 7020.
Eldridge, M.D., Murray, C.W., Auton, T.R., Paolini, G.V. and Mee, R.P., J. Comput.-Aided Mol. Design, 11 (1997) 425.
DeWitte, R.S. and Shakhnovich, E.I., J. Am. Chem. Soc., 118 (1996) 11733.
Muegge, I. and Martin, Y.C., J. Med. Chem., 42 (1999) 791.
Böhm, H.-J., J. Comput.-Aided Mol. Design, 8 (1998) 243.
Weiner, S.J., Kollman, P.A., Case, D.A., Singh, U.C., Ghio, C., Alagona, G., Profeta Jr., S. and Weiner, P., J. Am. Chem. Soc., 106 (1984) 765.
Weiner, S.J., Kollman, P.A., Nguyen, D.T. and Case, D.A., J. Comput. Chem., 7 (1986) 230.
Muegge, I., Martin, Y.C., Hajduk, P.J. and Fesik, S.W., J. Med. Chem., 42 (1999) 2498.
Muegge, I., Med. Chem. Res., 9 (1999) 490.
Birkedal-Hansen, H., Moore, W.G.I., Bodden, M.K., Windsor, L.J., Birkedal-Hansen, B., DeCarlo, A. and Engler, J.A., Crit. Rev. Oral Biol. Med., 4 (1993) 197.
Woessner, J.F.J., FASEB J., 5 (1991) 2145.
Dhanaraj, V., Ye, Q.Z., Johnson, L.L., Hupe, D.J., Ortwine, D.F., Dunbar, J.B., Rubin, J.R., Pavolvski, A., Humblet, C. and Blundell, T.L., Structure, 4 (1996) 466.
Stockman, B.J., Watson, D.J., Gates, J.A., Scahill, T.A., Kloosterman, D.A., Miszak, S.A., Jacobsen, E.J., Belonga, K.L., Mitchell, M.A., Mao, B., Petke, J.D., Goodman, L. and Powers, E.A., Protein Sci., 7 (1998) 2281.
Marcy, A.I., Eiberger, L.L., Harrison, R., Chan, H.K., Hutchinson, N.L., Hagman, W.K., Cameron, P.M., Boulton, D.A. and Hermes, J.D., Biochemistry, 30 (1991) 6476.
Howard, A.J., Nielsen, C. and Xuong, N.H., Methods Enzymol., 114 (1985) 452.
Furey, W.F. and Swaminathan, S., Methods Enzymol., 227 (1997) 590.
Wang, B.C., Methods Enzymol., 114 (1985) 90.
Jones, T.A., J. Appl. Crystallogr., 115 (1975) 157.
Brünger, A.T., X-PLOR Manual, Version 3.1, Yale University Press, New Haven, CT, 1992.
Sippl, M.J., J. Mol. Biol., 213 (1990) 859.
Sippl, M.J., J. Comput.-Aided Mol. Design, 7 (1993) 473.
Sippl, M.J., Ortner, M., Jaritz, M., Lackner, P. and Flöckner, H., Folding Design, 1 (1996) 289.
Warshel, A., Papazyan, A. and Muegge, I., J. Biol. Inorg. Chem., 2 (1997) 143.
Muegge, I., Qi, X.P., Wand, A.J., Chu, Z.T. and Warshel, A., J. Phys. Chem. B, 101 (1997) 825.
Muegge, I., Tao, H. and Warshel, A., Protein Eng., 10 (1997) 1363.
Muegge, I., Perspect. Drug Discov. Des., accepted for publication.
Gohlke, H., Hendlich, M. and Klebe, G., J. Mol. Biol., 295 (2000) 337.
Walters, W.P., Stahl, M.T. and Murcko, M.A., Drug Discov. Today, 3 (1998) 160.
Kluender, H.C.E., Dixon, B.R., VanZandt, M.C., Willhelm, S.M., Wolanin, D.J. and Wood, J.E., U.S. Patent No. 5,861,428, 1999.
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Ha, S., Andreani, R., Robbins, A. et al. Evaluation of docking/scoring approaches: A comparative study based on MMP3 inhibitors. J Comput Aided Mol Des 14, 435–448 (2000). https://doi.org/10.1023/A:1008137707965
Issue Date:
DOI: https://doi.org/10.1023/A:1008137707965