Abstract
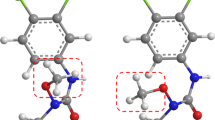
CoMFA analysis, a widely used 3D-QSAR method, has limitations to handle a set of SAR data containing diverse conformational flexibility since it does not explicitly include the conformational entropic effects into the analysis. Here, we present an attempt to incorporate the conformational entropy effects of a molecule into a 3D-QSAR analysis. Our attempt is based on the assumption that the conformational entropic loss of a ligand upon making a ligand-receptor complex is small if the ligand in an unbound state has a conformational propensity to adopt an active conformation in a complex state. For a QSAR analysis, this assumption was interpreted as follows: a potent ligand should have a higher conformational propensity to adopt an `active-conformation'-like structure in an unbound state than an inactive one. The conformational propensity value was defined as the populational ratio, Nactive/Nstable, of the number of energetically stable conformers, Nstable, to the number of `active-conformation'-like structures, Nactive. The latter number was calculated by counting the number of conformers that satisfied the structural parameters deduced from the active conformation. A set of SAR data of imidazoleglycerol phosphate dehydratase inhibitors containing 20 molecules with different conformational flexibility was used as a training set for developing a 3D structure-activity relationship by a CoMFA analysis with the conformational propensity value. This resulted in a cross-validated squared correlation coefficient of the CoMFA model with the conformational propensity value (R 2 cross = 0.640) higher than that of the standard CoMFA model (R 2 cross = 0.431). Then we evaluated the quality of the CoMFA models by predicting the inhibitory activity for a new molecule.
Similar content being viewed by others
References
Bohacek, R.S., McMartin, C. and Guida, W.C., Med. Res. Rev., 16 (1996) 3.
Kubinyi, H., Drug Discov. Today, 2 (1997) 457.
Kubinyi, H., Drug Discov. Today, 2 (1997) 538.
Kubinyi, H. (Ed.) 3D-QSAR in Drug Design: Theory, Methods and Applications, ESCOM, Leiden, 1993.
Stahle, L. and Wold, S., Prog. Med. Chem., 25 (1988) 292.
Cramer, R.D., III, J. Am. Chem. Soc., 110 (1988) 5959.
Marshall, G.R., Barry, C.D., Bosshard, H.E., Dammkoehler, R.A. and Dunn, D.A., in Olsen, E.C. and Christoffersen, E. (Eds.) Computer Assisted Drug Design, American Chemical Society, Washington, DC, 1979, pp. 87–112.
Klebe, G., In Kubinyi, H. (Ed.) 3D-QSAR in Drug Design: Theory, Methods and Applications, ESCOM, Leiden, 1993, pp. 173–199.
Martin, Y.C., Bures, M.G., Danaher, E.A., DeLazzer, J., Lico, I. and Pavlik, P.A., J. Comput.-Aided Mol. Design, 7 (1993) 83.
Sprague, P. W., Perspect. Drug Discov. Des., 3 (1995) 1.
Lemmen, C. and Lengauer, T., J. Comput.-Aided Mol. Design, 11 (1997) 357.
Lemmen, C., Lengauer, T. and Klebe, G., J. Med. Chem., 41 (1998) 4502.
Klebe, G. and Abraham, U., J. Med. Chem., 36 (1993) 70.
DePriest, S.A., Mayer, D., Naylor, C.B. and Marshall, G.R., J. Am. Chem. Soc., 115 (1993) 5372.
Kellogg, G.E., Semus, S.F. and Abraham, D.J., J. Comput.-Aided Mol. Design, 5 (1991) 545.
Gaillard, P., Carrupt, P.-A., Testa, B. and Boudon, A., J. Comput.-Aided Mol. Design, 8 (1994) 83.
Masuda, T., Nakamura, K., Jikihara, T., Kasuya, F., Igarashi, K., Fukui, M., Takagi, T. and Fujiwara, H., Quant. Struct.-Act. Relat., 15 (1996) 194.
Ghose, A.K., Viswanadhan, V.N. and Wendoloski, J.J., J. Phys. Chem. A, 102 (1998) 3762.
Kim, K.H., Quant. Struct.-Act. Relat., 12 (1993) 232.
Koehler, M.G. and Hopfinger, A.J., Polymer, 30 (1989) 116.
Tokarski, J.S. and Hopfinger, A.J., J. Chem. Inf. Comput. Sci., 37 (1997) 792.
Hopfinger, A.J., Wang, S., Tokarski, J.S., Jin, B., Albuquerque, M., Madhav, P.J. and Duraiswami, C., J. Am. Chem. Soc., 119 (1997) 10509.
Ames, B.N., J. Biol. Chem., 228 (1957) 131.
Hawkes, T.R., Cox, J.M., Barnes, N.J., Beautement, K., Edwards, L.S., Kipps, M.R., Langford, M.P., Lewis, T., Ridley, S.M. and Thomas, P.G., Brighton Crop Protection Conference (Weeds), 1993, pp. 739–744.
Mori, I., Fonne-Pfister, R., Matsunaga, S., Tada, S., Kimura, Y., Iwasaki, G., Mano, J., Hatano, M., Nakano, T., Koizumi, S., Scheidegger, A., Hayakawa, K. and Ohta, D., Plant Physiol., 107 (1995) 719.
Lindell, S.D., Earnshaw, C.G., Wright, B.J., Carver, D.S., O'Mahony, M.J. and Saville-Stones, E.A., Bioorg. Med. Chem. Lett., 6 (1996) 547.
Mori, I., Iwasaki, G., Kimura, Y., Matsunaga, S., Ogawa, A., Nakano, T., Buser, H.-P., Hatano, M., Tada, S. and Hayakawa, K., J. Am. Chem. Soc., 117 (1995) 4411.
QUANTA v. 3.3.1., Molecular Simulation Inc., Burlington, MA, 1994.
Brooks, B.R., Bruccoleri, R.E., Olafson, B.D., States, D.J., Swaminathan, S. and Karplus, M., J. Comput. Chem., 4 (1983) 187.
Stewart, J.J.P., J. Comput. Chem., 10 (1989) 209.
SPARTAN v. 4.0.2.,Wavefunction Co., Irvine, CA, 1995.
Chang, G., Guida, W.C. and Still, W.C., J. Am. Chem. Soc., 111 (1989) 4379.
Guida, W.C., Bohacek, R.S. and Erion, M.D., J. Comput. Chem., 13 (1992) 214.
SYBYL v. 6.0.2., Tripos Inc., St. Louis, MO, 1994.
Cramer III, R.D., DePriest, S.A., Patterson, D.E. and Hecht, P., in Kubinyi, H. (Ed.) 3D-QSAR in Drug Design: Theory, Methods and Applications, ESCOM, Leiden, 1993, pp. 443–485.
Wold, S., Technometrics, 20 (1978) 397.
SYBYL 6.0 manual: Ligand-Based Design, Tripos Inc., St. Louis, MO, 1994, pp. 195–262.
Leo, A.J., CLOGP version 3.63, Daylight Chemical Information Systems, Irvine, CA, 1991.
Bostrom, J., Norrby, P.-O. and Liljefors, T., J. Comput.-Aided Mol. Design, 12 (1998) 383.
Gohda, K., Kimura, Y., Mori, I., Ohta, D. and Kikuchi, T., Biochim. Biophys. Acta, 1385 (1998) 107.
QUANTA 3.3 manual: Simulation, search, and analysis, Molecular Simulation Inc., Burlington, MA, 1994, pp. 185–228.
Clark, M. and Cramer, R.D., III, Quant. Struct.-Act. Relat., 12 (1993) 137.
Epps, D.E., Cheney, J., Schostarez, H., Sawyer, T.K., Prairie, M., Krueger, W.C. and Mandel, F., J. Med. Chem., 33 (1990) 2080.
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Gohda, K., Mori, I., Ohta, D. et al. A CoMFA analysis with conformational propensity: An attempt to analyze the SAR of a set of molecules with different conformational flexibility using a 3D-QSAR method. J Comput Aided Mol Des 14, 265–275 (2000). https://doi.org/10.1023/A:1008193217627
Issue Date:
DOI: https://doi.org/10.1023/A:1008193217627