Abstract
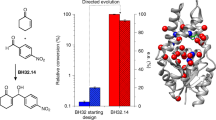
The combination of computational design and laboratory evolution is a powerful and potentially versatile strategy for the development of enzymes with new functions1,2,3,4. However, the limited functionality presented by the genetic code restricts the range of catalytic mechanisms that are accessible in designed active sites. Inspired by mechanistic strategies from small-molecule organocatalysis5, here we report the generation of a hydrolytic enzyme that uses Nδ-methylhistidine as a non-canonical catalytic nucleophile. Histidine methylation is essential for catalytic function because it prevents the formation of unreactive acyl-enzyme intermediates, which has been a long-standing challenge when using canonical nucleophiles in enzyme design6,7,8,9,10. Enzyme performance was optimized using directed evolution protocols adapted to an expanded genetic code, affording a biocatalyst capable of accelerating ester hydrolysis with greater than 9,000-fold increased efficiency over free Nδ-methylhistidine in solution. Crystallographic snapshots along the evolutionary trajectory highlight the catalytic devices that are responsible for this increase in efficiency. Nδ-methylhistidine can be considered to be a genetically encodable surrogate of the widely employed nucleophilic catalyst dimethylaminopyridine11, and its use will create opportunities to design and engineer enzymes for a wealth of valuable chemical transformations.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
Coordinates and structure factors have been deposited in the Protein Data Bank under accession numbers 6Q7N, 6Q7O, 6Q7P, 6Q7Q and 6Q7R. The authors declare that the data supporting the findings of this study are available within the paper and its Supplementary Information files. Source data are available from the corresponding author upon reasonable request.
References
Röthlisberger, D. et al. Kemp elimination catalysts by computational enzyme design. Nature 453, 190–195 (2008).
Siegel, J. B. et al. Computational design of an enzyme catalyst for a stereoselective bimolecular Diels–Alder reaction. Science 329, 309–313 (2010).
Blomberg, R. et al. Precision is essential for efficient catalysis in an evolved Kemp eliminase. Nature 503, 418–421 (2013).
Obexer, R. et al. Emergence of a catalytic tetrad during evolution of a highly active artificial aldolase. Nat. Chem. 9, 50–56 (2017).
Jencks, W. P., Salvesen, K. & Oakenfull, D. G. Reactions of acetylimidazole and acetylimidazolium ion with nucleophilic reagents. Mechanisms of catalysis. J. Am. Chem. Soc. 93, 188–194 (1971).
Bolon, D. N. & Mayo, S. L. Enzyme-like proteins by computational design. Proc. Natl Acad. Sci. USA 98, 14274–14279 (2001).
Richter, F. et al. Computational design of catalytic dyads and oxyanion holes for ester hydrolysis. J. Am. Chem. Soc. 134, 16197–16206 (2012).
Rajagopalan, S. et al. Design of activated serine-containing catalytic triads with atomic-level accuracy. Nat. Chem. Biol. 10, 386–391 (2014).
Moroz, Y. S. et al. New tricks for old proteins: single mutations in a nonenzymatic protein give rise to various enzymatic activities. J. Am. Chem. Soc. 137, 14905–14911 (2015).
Burton, A. J., Thomson, A. R., Dawson, W. M., Brady, R. L. & Woolfson, D. N. Installing hydrolytic activity into a completely de novo protein framework. Nat. Chem. 8, 837–844 (2016).
Wurz, R. P. Chiral dialkylaminopyridine catalysts in asymmetric synthesis. Chem. Rev. 107, 5570–5595 (2007).
Studer, S. et al. Evolution of a highly active and enantiospecific metalloenzyme from short peptides. Science 362, 1285–1288 (2018).
Brady, L. et al. A serine protease triad forms the catalytic centre of a triacylglycerol lipase. Nature 343, 767–770 (1990).
Smith, A. J. T. et al. Structural reorganization and preorganization in enzyme active sites: comparisons of experimental and theoretically ideal active site geometries in the multistep serine esterase reaction cycle. J. Am. Chem. Soc. 130, 15361–15373 (2008).
Buller, A. R. & Townsend, C. A. Intrinsic evolutionary constraints on protease structure, enzyme acylation, and the identity of the catalytic triad. Proc. Natl Acad. Sci. USA 110, E653–E661 (2013).
Breslow, R. & Dong, S. D. Biomimetic reactions catalyzed by cyclodextrins and their derivatives. Chem. Rev. 98, 1997–2012 (1998).
Bjelic, S. et al. Computational design of enone-binding proteins with catalytic activity for the Morita–Baylis–Hillman reaction. ACS Chem. Biol. 8, 749–757 (2013).
Xiao, H. et al. Genetic incorporation of histidine derivatives using an engineered pyrrolysyl-tRNA synthetase. ACS Chem. Biol. 9, 1092–1096 (2014).
Bornscheuer, U. T. et al. Engineering the third wave of biocatalysis. Nature 485, 185–194 (2012).
Arnold, F. H. Directed evolution: bringing new chemistry to life. Angew. Chem. Int. Ed. 57, 4143–4148 (2018).
Reetz, M. T., Bocola, M., Carballeira, J. D., Zha, D. & Vogel, A. Expanding the range of substrate acceptance of enzymes: combinatorial active-site saturation test. Angew. Chem. Int. Ed. 44, 4192–4196 (2005).
Kawai, F. et al. A novel Ca2+-activated, thermostabilized polyesterase capable of hydrolyzing polyethylene terephthalate from Saccharomonospora viridis AHK190. Appl. Microbiol. Biotechnol. 98, 10053–10064 (2014).
Griffin, J. L. et al. Near attack conformers dominate β-phosphoglucomutase complexes where geometry and charge distribution reflect those of substrate. Proc. Natl Acad. Sci. USA 109, 6910–6915 (2012).
Liu, C. C. & Schultz, P. G. Adding new chemistries to the genetic code. Annu. Rev. Biochem. 79, 413–444 (2010).
Chin, J. W. Expanding and reprogramming the genetic code. Nature 550, 53–60 (2017).
Ugwumba, I. N. et al. Improving a natural enzyme activity through incorporation of unnatural amino acids. J. Am. Chem. Soc. 133, 326–333 (2011).
Agostini, F. et al. Biocatalysis with unnatural amino acids: enzymology meets xenobiology. Angew. Chem. Int. Ed. 56, 9680–9703 (2017).
Windle, C. L. et al. Extending enzyme molecular recognition with an expanded amino acid alphabet. Proc. Natl Acad. Sci. USA 114, 2610–2615 (2017).
Yang, H. et al. Evolving artificial metalloenzymes via random mutagenesis. Nat. Chem. 10, 318–324 (2018).
Drienovská, I., Mayer, C., Dulson, C. & Roelfes, G. A designer enzyme for hydrazone and oxime formation featuring an unnatural catalytic aniline residue. Nat. Chem. 10, 946–952 (2018).
Lee, T. S. et al. BglBrick vectors and datasheets: A synthetic biology platform for gene expression. J. Biol. Eng. 5, 12 (2011).
Kille, S. et al. Reducing codon redundancy and screening effort of combinatorial protein libraries created by saturation mutagenesis. ACS Synth. Biol. 2, 83–92 (2013).
Yang, Y., Babiak, P. & Reymond, J. New monofunctionalized fluorescein derivatives for the efficient high-throughput screening of lipases and esterases in aqueous media. Helv. Chim. Acta 89, 404–415 (2006).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D 66, 213–221 (2010).
Acknowledgements
We acknowledge the Biotechnology and Biological Sciences Research Council (David Phillips Fellowship BB/M027023/1 to A.P.G) and the European Research Council (ERC Starter Grant, no. 757991 to A.P.G). We thank the Biotechnology and Biological Sciences Research Council and the Faculty of Science and Engineering (University of Manchester) for the award of an UKRI Innovation fellowship to S.L.L. We are grateful to Diamond Light Source for time on beamlines i03, i04 and i04-1 under proposals MX12788-50, MX17773-25, MX17773-34 and MX17773-52, and to Manchester SYNBIOCHEM Centre (BB/M017702/1) and the Future Biomanufacturing Hub (EP/S01778X/1) for access to their facilities and for guidance on automating directed-evolution workflows. We thank R. Spiess (Manchester Institute of Biotechnology) for acquiring protein mass spectra and R. Smithson for assistance with the chemical synthesis of activated esters. The pET29_BH32 plasmid was a gift from D. Baker.
Reviewer information
Nature thanks Uwe Bornscheuer, Adam Nelson and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Author information
Authors and Affiliations
Contributions
A.J.B. and S.L.L. carried out molecular biology, assay development, directed evolution, and protein production, purification and kinetic characterization. R.C., A.J.B. and S.L.L. carried out enzyme-inhibition experiments and protein crystallization. M.D. automated the directed-evolution workflow. A.F., M.O., M.D. and C.L. interpreted and analysed structural data. All authors discussed the results and participated in writing the manuscript. A.P.G. initiated and directed the research.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Kinetic characterization of BH32.
Michaelis–Menten plot showing the rate of His23 acylation (the ‘burst phase’) with varying concentrations of fluorescein 2-phenylacetate. Averaged initial rates were fitted to the Michaelis–Menten equation to derive rate constant kobs 1.3 ± 0.03 min−1 and the enzyme–substrate dissocation constant KS 30.9 ± 1.8 µM (R2 = 0.99). Data are mean ± s.d. of measurements made in triplicate.
Extended Data Fig. 2 Evolutionary strategy used to generate OE1.3 and OE1.4.
Structures showing the amino acid positions randomized during each round of evolution. The Me-His residue is shown in ball-and-stick representation and targeted residues are represented as spheres. a, The round 1 library was prepared by randomizing active-site residues in pairs Ala19/Ser22 (blue) Tyr45/Glu46 (orange) Tyr87/Trp88 (red) Met94/Ser95 (green) and Asp125/Gln128 (purple). b, The round 2 library was prepared by random mutagenesis of the entire gene. Variants with improved activity enabled the identification of ‘hot spots’, which were further interrogated using saturation mutagenesis. Residues Leu42/Glu46 (red) and Phe132/Leu133 (blue) were randomized simultaneously; all other positions were randomized individually (green). c, The round 3 library was prepared by saturation mutagenesis, using NNK degenerate codons to individually randomize 21 positions (see table). d, The round 4 library was prepared by saturation mutagenesis, using NNK degenerate codons to individually randomize 20 positions (see table). Libraries generated during rounds 1–3 were screened for activity towards fluorescein 2-phenylacetate. The round 4 library was screened for activity towards fluorescein (R)-2-phenylpropanoate.
Extended Data Fig. 3 Kinetic characterization of OE1, OE1 variants and CUT190.
a–f, Michaelis–Menten plots of fluorescein 2-phenylacetate hydrolysis catalysed by OE1 (a), OE1.1 (b), OE1.2 (c), OE1.3 (d), OE1.3(N46E) (e) and OE1.3(Y45F) (f). g, A Hill plot of fluorescein 2-phenylacetate hydrolysis catalysed by CUT190, n = 2.3 ± 0.4. Data are mean ± s.d. of measurements made in triplicate. h, A table summarizing the kinetic parameters of OE1 and its variants, and the cutinase variant Ser226Pro/Arg228Ser (CUT190).
Extended Data Fig. 4 Kinetic characterization of OE1.3 and OE1.4 towards the hydrolysis of (R)- and (S)- fluorescein 2-phenylpropanoate.
a, Michaelis–Menten plots of fluorescein (R)-2-phenylpropanoate and fluorescein (S)-2-phenylpropanoate hydrolysis (shown in black and red, respectively), catalysed by OE1.3. The averaged initial rates were fitted to the Michaelis–Menten equation using Origin software. b, Michaelis–Menten plots of fluorescein (R)-2-phenylpropanoate and fluorescein (S)-2-phenylpropanoate hydrolysis (shown in black and red, respectively) catalysed by OE1.4. The averaged initial rates were fitted to the equation for Michaelis–Menten with substrate inhibition using Origin software. Data are mean ± s.d. of measurements made in triplicate. c, Table summarizing the kinetic parameters for the hydrolysis of both enantiomers of fluorescein 2-phenylpropanoate catalysed by OE1.3 and OE1.4. Data are mean ± s.d. of measurements made in triplicate.
Extended Data Fig. 5 Rate constants of the hydrolysis of fluorescein 2-phenylacetate catalysed by small-molecule nucleophilic catalysts.
a–c, Linear plots showing the rate of hydrolysis of fluorescein 2-phenylacetate catalysed by 3-methylhistidine (Me-His) kMe-His = 0.35 M−1 s−1, R2 = 0.99) (a), dimethylaminopyridine (kDMAP = 1.13 M−1 s−1, R2 = 0.99) (b) and N-methylimidazole (kNMI = 1.16 M−1 s−1, R2 = 0.99) (c). d, Linear plot showing the rate of uncatalysed fluorescein 2-phenylacetate hydrolysis (kobs = 5.9 × 10−4 min−1, R2 = 0.98). Data are mean ± s.d. of measurements made in triplicate.
Extended Data Fig. 6 Comparison of the ester hydrolysis reaction catalysed by OE1.3 and OE1.3(C186A/C212A).
Time course of the hydrolysis of fluorescein 2-phenylacetate (100 µM) catalysed by OE1.3 (purple) and OE1.3(C186A/C212A) (black) (0.1 µM) in PBS pH 7.4, 22 °C, showing that the Cys186Ala and Cys212Ala mutations have a negligible effect on catalytic activity.
Extended Data Fig. 7 Structural characterization of inhibited BH32.
A ball-and-stick representation of H23 from BH32 inhibited with 2-bromoacetophenone, coloured by atom type with H23 carbon atoms in yellow and acetophenone carbons in white. Clear FEM electron density (blue, contoured at 1σ) extends between the Nε of H23 and acetophenone.
Extended Data Fig. 8 Structural characterization of inhibited BH32, OE1.2 and OE1.3.
A global superposition of BH32-inhibited (purple), OE1.2-inhibited (blue) and OE1.3-inhibited (green) structures performed using ICM Pro. The r.m.s.d. values (backbone atoms), derived from the global superposition are calculated separately for the core and cap domains and are reported in the table. BH32-inhibited and OE1.2-inhibited structures retain similar domain orientations, whereas OE1.3 undergoes a reorientation of the cap domain upon inhibition with 2-bromoacetophenone.
Supplementary information
Supplementary Information
This file contains the DNA and protein sequence of the most active variant OE1.3, 1H and 13C NMR spectra and X-ray crystallography tables
Rights and permissions
About this article
Cite this article
Burke, A.J., Lovelock, S.L., Frese, A. et al. Design and evolution of an enzyme with a non-canonical organocatalytic mechanism. Nature 570, 219–223 (2019). https://doi.org/10.1038/s41586-019-1262-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-019-1262-8
This article is cited by
-
A non-canonical nucleophile unlocks a new mechanistic pathway in a designed enzyme
Nature Communications (2024)
-
A designed photoenzyme for enantioselective [2+2] cycloadditions
Nature (2022)
-
Enantioselective [2+2]-cycloadditions with triplet photoenzymes
Nature (2022)
-
Building enzymes from scratch
Nature Chemistry (2022)
-
The road to fully programmable protein catalysis
Nature (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.