Abstract
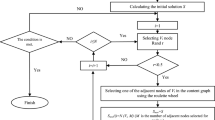
Graph algorithms are often used to analyze and interpret biological data. One of the widely used approaches is to solve the problem of identifying an active module, where a connected subgraph of a biological network is selected, which best reflects the difference between two biological states being considered. In this work, we extend this approach to the case of a larger number of biological states and formulate the problem of joint clustering in graph and correlation spaces. To solve this problem, an iterative method is proposed, which takes as the input the graph \(G\) and the matrix \(X\), in which the rows correspond to vertices of the graph. As the output, the algorithm generates a set of subgraphs of graph \(G\) so that each subgraph is connected and the rows corresponding to its vertices have a high pairwise correlation. The efficiency of the method is confirmed by an experimental study using simulated data.







Similar content being viewed by others
REFERENCES
Mitra, K., Carvunis, A.R., Ramesh, S.K., and Ideker, T., Integrative approaches for finding modular structure in biological networks, Nat. Rev. Genet., 2013, vol. 14, no. 10, pp. 719–732.
Rossin, E.J., et al., Proteins encoded in genomic regions associated with immune-mediated disease physically interact and suggest underlying biology, PLoS Genet., 2011, vol. 7, no. 1, e1001273.
Jha, A.K., Huang, S.C., Sergushichev, A., Lampropoulou, V., Ivanova, Y., Loginicheva, E., Chmielewski, K., Stewart, K.M., Ashall, J., Everts, B., Pearce, E.J., Driggers, E.M., and Artyomov, M.N., Network integration of parallel metabolic and transcriptional data reveals metabolic modules that regulate macrophage polarization, Immunity, 2015, vol. 42, no. 3, pp. 419–430.
Leiserson, M.D., et al., Pan-cancer network analysis identifies combinations of rare somatic mutations across pathways and protein complexes, Nat. Genet., 2015, vol. 47, no. 2, pp. 106–114.
Ideker, T., Ozier, O., Schwikowski, B., and Siegel, A.F., Discovering regulatory and signalling circuits in molecular interaction networks, Bioinformatics (Oxford, Engl.), 2002, vol. 18, suppl. 1, pp. S233–S240.
Dirich, M.T., Klau, G.W., Rosenwald, A., Dandekar, T., and Müller, T., Identifying functional modules in protein-protein interaction networks: An integrated exact approach, Bioinformatics (Oxford, Engl.), 2008, vol. 24, no. 13, pp. i223–i231. https://doi.org/10.1093/bioinformatics/btn161
Artyomov, M.N., Sergushichev, A., and Schilling, J.D., Integrating immunometabolism and macrophage diversity, Semin. Immunol., 2016, vol. 28, no. 5, pp. 417–424.
Loboda, A.A., Artyomov, M.N., and Sergushichev, A.A., Solving generalized maximum-weight connected subgraph problem for network enrichment analysis, Algorithms in Bioinformatics: 16th International Workshop, WABI 2016, Aarhus, Denmark, August 22–24, 2016. Proceedings, Cham: Springer Int. Publ., 2016, pp. 210–221.
Álvarez-Miranda, E.A. and Sinnl, M., A relax-and-cut framework for large-scale maximum weight connected subgraph problems, Comput. Oper. Res., 2017, vol. 87, pp. 63–82.
Langfelder, P. and Horvath, S., WGCNA: An R package for weighted correlation network analysis, BMC Bioinf., 2008, vol. 9, p. 559.
Funding
This study was funded by the Government of Russian Federation, project no. 08-08.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
The authors declare that they have no conflicts on interest.
Additional information
Translated by K. Lazarev
About this article
Cite this article
Gainullina, A.N., Shalyto, A.A. & Sergushichev, A.A. Method for Joint Clustering in Graph and Correlation Spaces. Aut. Control Comp. Sci. 55, 647–657 (2021). https://doi.org/10.3103/S0146411621070026
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.3103/S0146411621070026