Abstract
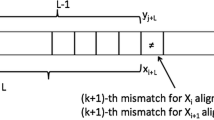
Fast and exact comparison of large genomic sequences remains a challenging task in biosequence analysis. We consider the problem of finding all ε-matches between two sequences, i.e. all local alignments over a given length with an error rate of at most ε. We study this problem theoretically, giving an efficient q-gram filter for solving it. Two applications of the filter are also discussed, in particular genomic sequence assembly and blast-like sequence comparison. Our results show that the method is 25 times faster than blast, while not being heuristic.
Preview
Unable to display preview. Download preview PDF.
Similar content being viewed by others
References
Altschul, S.F., Gish, W., Miller, W., Myers, E.W., Lipman, D.J.: Basic local alignment search tool. J. Mol. Biol. 215(3), 403–410 (1990)
Altschul, S.F., Madden, T.L., Schäffer, A.A., Zhang, J., Zhang, Z., Miller, W., Lipman, D.J.: Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25(17), 3389–3402 (1997)
Burkhardt, S., Crauser, A., Ferragina, P., Lenhof, H.-P., Rivals, E., Vingron, M.: q-gram based database searching using a suffix array. In: Proc. of the 3rd Annu. Int. Conf. on Computational Molecular Biology (RECOMB 1999), pp. 77–83 (1999)
Burkhardt, S., Kärkkäinen, J.: Better filtering with gapped q-grams. In: Amir, A., Landau, G.M. (eds.) CPM 2001. LNCS, vol. 2089, pp. 73–85. Springer, Heidelberg (2001)
Califano, A., Rigoutsos, I.: FLASH: a fast look-up algorithm for string homology. In: Proc. of the 1st Int. Conf. on Intelligent Systems for Molecular Biology (ISMB 1993), pp. 56–64 (1993)
Chang, W.I., Lawler, E.L.: Sublinear expected time approximate string matching and biological applications. Algorithmica 12(4/5), 327–344 (1994)
Eppstein, D., Galil, Z., Giancarlo, R., Italiano, G.F.: Sparse dynamic programming I: linear cost functions. J. ACM 39(3), 519–545 (1992)
Gusfield, D.: Algorithms on Strings, Trees and Sequences: Computer Science and Computational Biology. Cambridge University Press, Cambridge (1997)
Jokinen, P., Ukkonen, E.: Two algorithms for approximate string matching in static texts. In: Tarlecki, A. (ed.) MFCS 1991. LNCS, vol. 520, pp. 240–248. Springer, Heidelberg (1991)
Kent, W.J.: BLAT – the BLAST-like alignment tool. Genome Res. 12(4), 656–664 (2002)
Li, M., Ma, B., Kisman, D., Tromp, J.: PatternHunter II: Highly Sensitive and Fast Homology Search. In: Proc. of the 14th Annu. Int. Conf. on Genome Informatics (GIW 2003), pp. 164–175 (2003)
Ma, B., Tromp, J., Li, M.: PatternHunter – faster and more sensitive homology search. Bioinformatics 18, 440–445 (2002)
Myers, E.: A sublinear algorithm for approximate keyword searching. Algorithmica 12(4/5), 345–374 (1994)
Myers, E.: A fast bit-vector algorithm for approximate string matching based on dynamic programming. J. ACM 46(3), 539–553 (1999)
Myers, E., Durbin, R.: A table-driven, full-sensitivity similarity search algorithm. J. Comp. Bio. 10(2), 103–118 (2003)
Ning, Z., Cox, A.J., Mullikin, J.C.: SSAHA: A fast search method for large DNA databases. Genome Res. 11(10), 1725–1729 (2001)
Pearson, W.R., Lipman, D.J.: Improved tools for biological sequence comparison. Proc. Natl. Acad. Sci. USA 85, 2444–2448 (1988)
Pearson, W.R.: Searching protein sequence libraries: Comparison of the sensitivity and selectivity of the Smith-Waterman and FASTA algorithms. Genomics 11, 635–650 (1991)
Smith, T.F., Waterman, M.S.: Identification of common molecular subsequences. J. Mol. Biol. 147(1), 195–197 (1981)
Ukkonen, E.: Approximate string-matching with q-grams and maximal matches. Theor. Comput. Sci. 92(1), 191–211 (1992)
Zhang, Z., Berman, P., Miller, W.: Alignments without low-scoring regions. In: Proc. of the 2nd Annu. Int. Conf. on Computational Molecular Biology (RECOMB 1998), pp. 294–301 (1998)
ftp://ftp.ncbi.nlm.nih.gov/blast/db/FASTA/est_human,mouse.gz
Author information
Authors and Affiliations
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2005 Springer-Verlag Berlin Heidelberg
About this paper
Cite this paper
Rasmussen, K.R., Stoye, J., Myers, E.W. (2005). Efficient q-Gram Filters for Finding All ε-Matches over a Given Length. In: Miyano, S., Mesirov, J., Kasif, S., Istrail, S., Pevzner, P.A., Waterman, M. (eds) Research in Computational Molecular Biology. RECOMB 2005. Lecture Notes in Computer Science(), vol 3500. Springer, Berlin, Heidelberg. https://doi.org/10.1007/11415770_15
Download citation
DOI: https://doi.org/10.1007/11415770_15
Publisher Name: Springer, Berlin, Heidelberg
Print ISBN: 978-3-540-25866-7
Online ISBN: 978-3-540-31950-4
eBook Packages: Computer ScienceComputer Science (R0)