Abstract
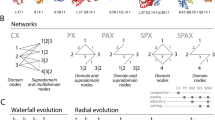
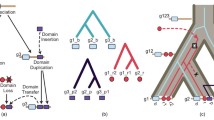
We study properties of multidomain proteins from a graph theoretical perspective. In particular, we demonstrate connections between properties of the domain overlap graph and certain variants of Dollo parsimony models. We apply our graph theoretical results to address several interrelated questions: do proteins acquire new domains infrequently, or often enough that the same combinations of domains will be created repeatedly through independent events? Once domain architectures are created, do they persist? In other words, is the existence of ancestral proteins with domain compositions not observed in contemporary proteins unlikely? Our experimental results indicate that independent merges of domain pairs are not uncommon in large superfamilies.
Preview
Unable to display preview. Download preview PDF.
Similar content being viewed by others
References
Apic, G., Gough, J., Teichmann, S.A.: Domain combinations in archaeal, eubacterial and eukaryotic proteomes. J. Mol. Biol. 310, 311–325 (2001)
Apic, G., Huber, W., Teichmann, S.A.: Multi-domain protein families and domain pairs: Comparison with known structures and a random model of domain recombination. J. Struc. Func. Genomics 4, 67–78 (2003)
Barabasi, A.-L., Albert, R.: Emergence of scaling in random networks. Science 286, 509–512 (1999)
Bashton, M., Chothia, C.: The geometry of domain combination in proteins. J. Mol. Biol. 315, 927–939 (2002)
Bateman, A., Birney, E., Durbin, R., Eddy, S.R., Howe, K.L., Sonnhammer, E.L.: The Pfam protein families database. Nucleic Acids Res. 28(1), 263–266 (2000)
Boeckmann, B., Bairoch, A., Apweiler, R., Blatter, M.-C., Estreicher, A., Gasteiger, E., Martin, M.J., Michoud, K., O’Donovan, C., Phan, I., Pilbout, S., Schneider, M.: The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res. 31, 365–370 (2003)
Bollobas, B.: Random Graph Theory. Cambridge University Press, Cambridge (2001)
Cheek, S., Zhang, H., Grishin, N.V.: Sequence and structure classification of kinases. J. Mol. Biol. 320(4), 855–881 (2002)
Danzer, L., Grunbaum, B., Klee, V.: Helly’s theorem and its relatives. Convexity, AMS 7, 101–180 (1963)
Day, W.H.E., Johnson, D., Sankoff, D.: The computational complexity of inferring rooted phylogenies by parsimony. Mathematical Biosciences 81, 33–42 (1986)
Gusfield, D.: Efficient methods for inferring evolutionary history. Networks 21, 19–28 (1991)
Felsenstein, J.: Inferring Phylogenies. Sinauer Associates (2004)
Gavril, F.: The intersection graphs of subtrees in trees are exactly the chordal graphs. J. Comb. Theory (B) 16, 47–56 (1974)
Geer, L.Y., Domrachev, M., Lipman, D.J., Bryant, S.H.: CDART: protein homology by domain architecture. Genome Res. 12(10), 1619–1623 (2002)
Gerstein, M.: How representative are the known structures of the proteins in a complete genome? A comprehensive structural census. Fold des. 3, 497–512 (1998)
Golumbic, M.: Algorithmic Graph Theory and Perfect Graphs. Academic Press, New York (1980)
Gu, J., Gu, X.: Natural history and functional divergence of protein tyrosine kinases. Gene. 317, 49–57 (2003)
Hanks, S.K.: Genomic analysis of the eukaryotic protein kinase superfamily: a perspective. Genome Biol. 4(5), 111 (2003)
Heger, A., Holm, L.: Exhaustive enumeration of protein domain families. J. Mol. Biol. 328, 749–767 (2003)
Yanai, I., Wolf, Y.I., Koonin, E.V.: Evolution of gene fusions: horizontal transfer versus independent events. Genome Biol. 3 (2002), research:0024
Farris, J.S.: Phylogenetic analysis under Dollo’s law. Systematic Zoology 26(1), 77–88 (1977)
Krause, A., Stoye, J., Vingron, M.: The SYSTERS protein sequence cluster set. Nucleic Acids Res. 28(1), 270–272 (2000)
Kummerfeld, S., Vogel, C., Madera, M., Teichmann, S.: Evolution of multi-domain proteins by gene fusion and fission. In: ISMB 2004 (2004)
Letunic, I., Goodstadt, L., Dickens, N.J., Doerks, T., Schultz, J., Mott, R., Ciccarelli, F., Copley, R.R., Ponting, C.P., Bork, P.: Recent improvements to the SMART domain-based sequence annotation resource. Nucleic Acids Res. 31(1), 242–244 (2002)
Liu, Y., Gerstein, M., Engelman, D.M.: Evolutionary use of domain recombination: a distinction between membrane and soluble proteins. Proc. Natl. Acad. Sci. USA, 3495–3497 (2004)
Long, M.: Evolution of novel genes. Curr. Opin. Genet. Dev. 11(6), 673–680 (2001)
Patthy, L.: Genome evolution and the evolution of exon-shuffling–a review. Gene 238, 103–114 (1999)
Marcotte, E.M., Pellegrini, M., Ng, H.L., Rice, D.W., Yeates, T.O., Eisenberg, D.: Detecting protein function and protein-protein interactions from genome sequences. Science 285, 751–753 (1999)
Mehlhorn, K., Naher, S.: The LEDA Platform of Combinatorial and Geometric Computing. Cambridge University Press, Cambridge (1999)
Robinson, D.R., Wu, Y.M., Lin, S.F.: The protein tyrosine kinase family of the human genome. Oncogene 19(49), 5548–5558 (2000)
Teichmann, S.A., Park, J., Chothia, C.: Structural assignments to the mycoplasma genitalium proteins show extensive gene duplications and domain rearrangements (1998)
Snel, B., Bork, P., Huynen, M.: Genome evolution gene fusion versus gene fission. Trends Genet. 16, 9–11 (2002)
Wuchty, S.: Scale-free behavior in protein domain networks. Mol. Biol. Evol. 18, 1694–1702 (2001)
Yona, G., Linial, N., Linial, M.: Protomap: Automatic classification of protein sequences, a hierarchy of protein families, and local maps of the protein space. Proteins: Structure, Function and Genetics 37, 360–378 (1999)
Author information
Authors and Affiliations
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2005 Springer-Verlag Berlin Heidelberg
About this paper
Cite this paper
Przytycka, T., Davis, G., Song, N., Durand, D. (2005). Graph Theoretical Insights into Evolution of Multidomain Proteins. In: Miyano, S., Mesirov, J., Kasif, S., Istrail, S., Pevzner, P.A., Waterman, M. (eds) Research in Computational Molecular Biology. RECOMB 2005. Lecture Notes in Computer Science(), vol 3500. Springer, Berlin, Heidelberg. https://doi.org/10.1007/11415770_24
Download citation
DOI: https://doi.org/10.1007/11415770_24
Publisher Name: Springer, Berlin, Heidelberg
Print ISBN: 978-3-540-25866-7
Online ISBN: 978-3-540-31950-4
eBook Packages: Computer ScienceComputer Science (R0)