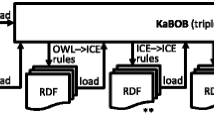
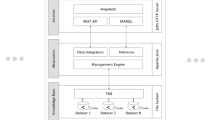
Abstract
We present INDUS (Intelligent Data Understanding System), a federated, query-centric system for knowledge acquisition from autonomous, distributed, semantically heterogeneous data sources that can be viewed (conceptually) as tables. INDUS employs ontologies and inter-ontology mappings, to enable a user or an application to view a collection of such data sources (regardless of location, internal structure and query interfaces) as though they were a collection of tables structured according to an ontology supplied by the user. This allows INDUS to answer user queries against distributed, semantically heterogeneous data sources without the need for a centralized data warehouse or a common global ontology. We used INDUS framework to design algorithms for learning probabilistic models (e.g., Naive Bayes models) for predicting GO functional classification of a protein based on training sequences that are distributed among SWISSPROT and MIPS data sources. Mappings such as EC2GO and MIPS2GO were used to resolve the semantic differences between these data sources when answering queries posed by the learning algorithms. Our results show that INDUS can be successfully used for integrative analysis of data from multiple sources needed for collaborative discovery in computational biology.
Preview
Unable to display preview. Download preview PDF.
Similar content being viewed by others
References
Andorf, C., Silvescu, A., Dobbs, D., Honavar, V.: Learning classifiers for assigning protein sequences to gene ontology functional families. In: Fifth International Conference on Knowledge Based Computer Systems (KBCS 2004), India (2004)
Ashburner, M., Ball, C., Blake, J., Botstein, D., Butler, H., Cherry, J., Davis, A., Dolinski, K., Dwight, S., Eppig, J., Harris, M., Hill, D., Issel-Tarver, L., Kasarskis, A., Lewis, S., Matese, J., Richardson, J., Ringwald, M., Rubin, G., Sherlock, G.: Gene ontology: tool for unification of biology. Nature Genetics 25(1), 25–29 (2000)
Baader, F., Nutt, W.: Basic description logics. In: Baader, F., Calvanese, D., McGuinness, D., Nardi, D., Patel-Schneider, P.F. (eds.) The Description Logic Handbook: Theory, Implementation, and Applications, pp. 43–95. Cambridge University Press, Cambridge (2003)
Bao, J., Honavar, V.: Collaborative ontology building with wiki@nt - a multi-agent based ontology building environment. In: McIlraith, S.A., Plexousakis, D., van Harmelen, F. (eds.) ISWC 2004. LNCS, vol. 3298. Springer, Heidelberg (2004)
Berners-Lee, T., Hendler, J., Lassila, O.: The Semantic Web. Scientific American (May 2001)
Bonatti, P., Deng, Y., Subrahmanian, V.: An ontology-extended relational algebra. In: Proceedings of the IEEE Conference on Information Integration and Reuse, pp. 192–199. IEEE Press, Los Alamitos (2003)
Borgida, A., Serafini, L.: Distributed description logics: Directed domain correspondences in federated information sources. In: Proceedings of the Intenational Conference on Cooperative Information Systems (2002)
Calvanese, D., Giacomo, G.D., Lenzerini, M.: A framework for ontology integration. In: Proceedings of the international semantic web working symposium, Stanford, USA, pp. 303–316 (2001)
Caragea, D., Pathak, J., Honavar, V.: Learning classifiers from semantically heterogeneous data. In: Proceedings of the International Conference on Ontologies, Databases, and Applications of Semantics for Large Scale Information Systems (2004)
Caragea, D., Silvescu, A., Honavar, V.: A framework for learning from distributed data using sufficient statistics and its application to learning decision trees. International Journal of Hybrid Intelligent Systems 1(2) (2004)
Chen, J., Chung, S., Wong, L.: The Kleisli query system as a backbone for bioinformatics data integration and analisis. Bioinformatics, 147–188 (2003)
Davidson, S., Crabtree, J., Brunk, B., Schug, J., Tannen, V., Overton, G., Stoeckert, C.: K2/Kleisli and GUS: experiments in integrated access to genomic data sources. IBM Journal 40(2) (2001)
Eckman, B.: A practitioner’s guide to data management and data integration in bioinformatics. Bioinformatics, 3–74 (2003)
Eckman, B., Hernndez, M., Ho, H., Naumann, F., Popa, L.: Schema mapping and data integration with clio (demo and poster). In: Proceedings of the International Conference on Intelligent Systems for Molecular Biology (ISMB 2002), Edmonton, Canada (2002)
Etzold, T., Harris, H., Beulah, S.: SRS: An integration platform for databanks and analysis tools in bioinformatics. Bioinformatics Managing Scientific Data, 35–74 (2003)
Fikes, R., Farquhar, A., Rice, J.: Tools for assembling modular ontologies. In: The Fourteenth National Conference on Artificial Intelligence (1997)
Gruber, T.: Ontolingua: A mechanism to support portable ontologies
Haas, L., Schwarz, P., Kodali, P., Kotlar, E., Rice, J., Swope, W.: DiscoveryLink: a system for integrated access to life sciences data sources. IBM System Journal 40(2) (2001)
Hull, R.: Managing semantic heterogeneity in databases: A theoretical perspective. In: PODS, Tucson, Arizona, pp. 51–61 (1997)
Kargupta, H., Chan, P.: Advances in Distributed and Parallel Knowledge Discovery. AAAI/MIT, Cambridge (2000)
Kementsietsidis, A., Arenas, M., Miller, R.J.: Mapping data in peer-to-peer systems: Semantics and algorithmic issues. In: Proceedings of the ACM SIGMOD International Conference on Management of Data, pp. 325–336 (2003)
Kosky, A., Chen, I., Markowitz, V., Szeto, E.: Exploring heterogeneous biological databases: Tools and applications. In: Schek, H.-J., Saltor, F., Ramos, I., Alonso, G. (eds.) EDBT 1998. LNCS, vol. 1377, p. 499. Springer, Heidelberg (1998)
Mitra, P., Wiederhold, G., Kersten, M.: A graph-oriented model for articulation of ontology interdependencies. In: Conference on Extending Database Technology, Konstanz, Germany (2000)
Noy, N.F., Fergerson, R.W., Musen, M.A.: The knowledge model of protege-2000: Combining interoperability and flexibility. In: Dieng, R., Corby, O. (eds.) EKAW 2000. LNCS (LNAI), vol. 1937, pp. 17–32. Springer, Heidelberg (2000)
Shaker, R., Mork, P., Brockenbrough, J.S., Donelson, L., Tarczy-Hornoch, P.: The biomediator system as a tool for integrating biologic databases on the web. In: Proceedings of the Workshop on Information Integration on the Web (held in conjunction with VLDB 2004), Toronto, ON (2004)
Smith, M., Welty, C., McGuinness, D.: OWL Web Ontology Language Guide. W3C Recommendation (2004)
Staab, S., Studer, R.: Handbook on Ontologies. In: International Handbooks on Information Systems. Springer, Heidelberg (2004)
Stevens, R., Goble, C., Paton, N., Becchofer, S., Ng, G., Baker, P., Bass, A.: Complex query formulation over diverse sources in tambis. Bioinformatics, 189–220 (2003)
Tannen, V., Davidson, S., Harker, S.: The information integration in K2. Bioinformatics, 225–248 (2003)
Author information
Authors and Affiliations
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2005 Springer-Verlag Berlin Heidelberg
About this paper
Cite this paper
Caragea, D. et al. (2005). Information Integration and Knowledge Acquisition from Semantically Heterogeneous Biological Data Sources. In: Ludäscher, B., Raschid, L. (eds) Data Integration in the Life Sciences. DILS 2005. Lecture Notes in Computer Science(), vol 3615. Springer, Berlin, Heidelberg. https://doi.org/10.1007/11530084_15
Download citation
DOI: https://doi.org/10.1007/11530084_15
Publisher Name: Springer, Berlin, Heidelberg
Print ISBN: 978-3-540-27967-9
Online ISBN: 978-3-540-31879-8
eBook Packages: Computer ScienceComputer Science (R0)