Abstract
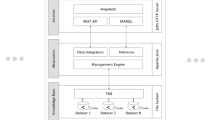
Biological sources integration has been addressed in several frameworks, considering both information sources incompatibilities and data representation heterogeneities. Most of these frameworks are mainly focused on coping with interoperability constraints among distributed databases that contain diverse types of biological data. In this paper, we propose an XML-based architecture that extends integration efforts from the distributed data sources domain to heterogeneous Bioinformatics tools of similar functionalities (“vertical integration”). The proposed architecture is based on the mediator/wrapper integration paradigm and a set of prescribed definitions that associates the capabilities and functional constraints of each analysis tool. The resulting XML-formatted information is further exploited by a visualization module that generates comparative views of the analysis outcome and a query mechanism that handles multiple information sources. The applicability of the proposed integration architecture and the information handling mechanisms was tested and substantiated on widely-known ab-initio gene finders that are publicly accessible through Web interfaces.
Preview
Unable to display preview. Download preview PDF.
Similar content being viewed by others
References
Stevens, R., Goble, C., Baker, P., Brass, A.: A Classification of Tasks in Bioinformatics. Bioinformatics 17(2), 180–188 (2001)
Wong, R.K., Shui, W.: Utilizing Multiple Bioinformatics Information Sources: An XML Databases Approach. In: Proceedings of IEEE Symposium on Bioinformatics and Bioengineering, pp. 73–80. IEEE Computer Society, Los Alamitos (2001)
Ng, S.-K., Wong, L.: Accomplishments and Challenges in Bioinformatics. IEEE IT Professional 6(1), 44–50 (2004)
Hernandez, T., Kambhampati, S.: Integration of Biological Sources: Current Systems and Challenges. ACM SIGMOD Record 33(3), 51–60 (2004)
Achard, F., Vaysseix, G., Barillot, E.: XML, Bioinformatics and Data Integration. Bioinformatics 17(2), 115–125 (2001)
World Wide Web Consortium: Extensible Markup Language (XML) 1.0 W3C Recommendation (3rd edn.) (2004), http://www.w3.org/TR/2004/REC-xml-20040204/
Entrez - Search and Retrieval System (2003), http://www.ncbi.nlm.nih.gov/Entrez
Wang, L., Riethoven, J.-J., Robinson, A.: XEMBL: Distributing EMBL Data in XML Format. Bioinformatics 18(8), 1147–1148 (2002)
Fenyö, D.: The Biopolymer Markup Language. Bioinformatics 15(4), 339–340 (1999)
Spellman, P.T., et al.: Design and Implementation of Microarray Gene Expression Markup Language (MAGE-ML). Genome Biology 3(9) (2002)
BSML, XML Data Standard for Genomics: The Bioinformatic Sequence Markup Language, http://www.bsml.org/
World Wide Web Consortium: XQuery 1.0: An XML Query Language (W3CWorking Draft) (2005), http://www.w3.org/TR/xquery/
Hammer, J., Schneider, M.: Genomics Algebra: A New, Integrating Data Model, Language, and Tool for Processing and Querying Genomic Information. In: Proceedings of the 2003 CIDR Conference (2003)
Tork Roth, M., Schwarz, P.: Don’t Scrap it, Wrap it! A Wrapper Architecture for Legacy Sources. In: Proceedings of the 23rd VLDB Conference Greece, pp. 266–275 (1997)
Koutkias, V., Malousi, A., Maglaveras, N.: Performing Ontology-driven Gene Prediction Queries in a Multi-Agent Environment. In: Barreiro, J.M., Martín-Sánchez, F., Maojo, V., Sanz, F. (eds.) ISBMDA 2004. LNCS, vol. 3337, pp. 378–387. Springer, Heidelberg (2004)
Davidson, S.B., Buneman, O.P., Crabtree, J., Tannen, V., Overton, G.C., Wong, L.: BioKleisli: Integrating Biomedical Data and Analysis Packages. In: Letovsky, S. (ed.) Bioinformatics: Databases and Systems, pp. 201–211. Kluwer Academic Publishers, Norwell (1999)
Baker, P., Brass, A., Bechhofer, S., Goble, C., Paton, N., Stevens, R.: TAMBIS: Transparent Access to Multiple Bioinformatics Information Sources. In: Proceedings of the 6th International Conference on Intelligent Systems for Molecular Biology (1998)
Haas, L., Schwarz, P., Kodali, P., Kotlar, E., Rice, J., Swope, W.: DiscoveryLink: A System for Integrated Access to Life Sciences Data Sources. IBM Systems Journal 40(2), 489–511 (2001)
Sujansky, W.: Heterogeneous Database Integration in Biomedicine. Journal of Biomedical Informatics 34, 285–298 (2001)
Aldana, J.F., Roldan, M., Navas, I., Perez, A.J., Trelles, O.: Integrating Biological Data Sources and Data Analysis Tools through Mediators. In: Proceedings of the 2004 ACM Symposium on Applied Computing Nicosia Cyprus, p. 127 (2004)
Zhang, M.Q.: Computational Prediction of Eukaryotic Protein-coding Genes. Nature 3, 698–710 (2002)
Mathé, C., Sagot, M.F., Schiex, T., Rouzé, P.: Current Methods of Gene Prediction, their Strengths and Weaknesses. Nucleic Acids Research 30, 4103–4117 (2002)
Rogic, S., Mackworth, A.K., Ouellette, F.: Evaluation of Gene-finding Programs on Mammalian Sequences. Genomic Research 11, 817–832 (2001)
Mironov, A.A., Fickett, J.W., Gelfand, M.S.: Frequent Alternative Splicing of Human Genes. Genome Research 9, 1288–1293 (1999)
Author information
Authors and Affiliations
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2005 Springer-Verlag Berlin Heidelberg
About this paper
Cite this paper
Malousi, A., Koutkias, V., Chouvarda, I., Maglaveras, N. (2005). Vertical Integration of Bioinformatics Tools and Information Processing on Analysis Outcome. In: Oliveira, J.L., Maojo, V., Martín-Sánchez, F., Pereira, A.S. (eds) Biological and Medical Data Analysis. ISBMDA 2005. Lecture Notes in Computer Science(), vol 3745. Springer, Berlin, Heidelberg. https://doi.org/10.1007/11573067_10
Download citation
DOI: https://doi.org/10.1007/11573067_10
Publisher Name: Springer, Berlin, Heidelberg
Print ISBN: 978-3-540-29674-4
Online ISBN: 978-3-540-31658-9
eBook Packages: Computer ScienceComputer Science (R0)