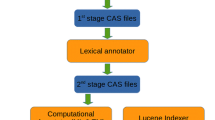
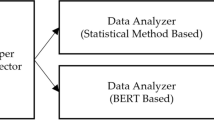
Abstract
The amount of information about the genome, transcriptome and proteome, forms a problem for the scientific community: how to find the right information in a reasonable amount of time. Most research aiming to solve this problem, however, concentrate on a certain organism or a very limited dataset. Complementary to those algorithms, we developed CONAN, a system which provides a full-scale approach, tailored to experimentalists, designed to combine several information extraction methods and connect the outcome of these methods to gather novel information. Its methods include tagging of gene/protein names, finding interaction and mutation data, tagging of biological concepts, linking to MeSH and Gene Ontology terms, which can all be found back by querying the system. We present a full-scale approach that will ultimately cover all of PubMed/MEDLINE. We show that this universality has no effect on quality: our system performs as well as existing systems.
Preview
Unable to display preview. Download preview PDF.
Similar content being viewed by others
References
Rebholz-Schuhmann, D., Kirsch, H., Couto, F.: Facts from text–is text mining ready to deliver? PLoS Biol. 3, e65 (2005)
Krallinger, M., Valencia, A.: Text-mining and information-retrieval services for molecular biology. Genome Biol 6, 224 (2005)
Canese, K., Jentsch, J., Myers, C.: The NCBI Handbook. National Center for Biotechnology Information (2003)
Ashburner, M., Ball, C., Blake, J., Botstein, D., Butler, H., Cherry, J., Davis, A., Dolinski, K., Dwight, S., Eppig, J., Harris, M., Hill, D., Issel-Tarver, L., Kasarskis, A., Lewis, S., Matese, J., Richardson, J., Ringwald, M., Rubin, G., Sherlock, G.: Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 25, 25–29 (2000)
Camon, E., Magrane, M., Barrell, D., Lee, V., Dimmer, E., Maslen, J., Binns, D., Harte, N., Lopez, R., Apweiler, R.: The Gene Ontology Annotation (GOA) Database: sharing knowledge in Uniprot with Gene Ontology. Nucleic Acids Res. 32, 262–266 (2004)
Apweiler, R., Bairoch, A., Wu, C.H., Barker, W.C., Boeckmann, B., Ferro, S., Gasteiger, R., Huang, H., Lopez, R., Magrane, M., Martin, M.J., Natale, D.A., O’Donovan, C., Redaschi, N., Yeh, L.S.: UniProt: the Universal Protein knowledgebase. Nucleic Acids Res. 32, D115–D119 (2004)
Birney, E., Andrews, T.D., Bevan, B., Caccamo, M., Chen, Y., Clarke, L., Coates, G., Cuff, J., Curwen, V., Cutts, T., Down, T., Eyras, E., Fernandez-Suarez, X., Gzane, P., Gibbins, B., Gilbert, J., Hammond, M., Hotz, H., Iyer, V., Jekosch, K., Kahari, A., Kasprzyk, A., Keefe, D., Keenan, S., Lehvaslaiho, H., McVicker, G., Melsopp, C., Meidl, P., Mongin, E., Pettett, R., Potter, S., Proctor, G., Rae, M., Searle, S., Slater, G., Smedley, D., Smith, J., Spooner, W., Stabenau, A., Stalker, J., Storey, R., Ureta-Vidal, A.: An Overview of Ensembl. Genome Res. 14, 925–928 (2004)
Krauthammer, M., Rzhetsky, A., Morozov, P., Friedman, C.: Using BLAST for identifying gene and protein names in journal articles. Gene. 259, 245–252 (2000)
Altschul, S., Gish, W., Miller, W., Myers, E., Lipman, D.: Basic local alignment search tool. J. Mol. Biol. 215, 403–410 (1990)
Bodenreider, O.: The Unified Medical Language System (UMLS): integrating biomedical terminology. Nucleic Acids Res. 32, 267–270 (2004)
Tanabe, L., Wilbur, W.: Tagging gene and protein names in biomedical text. Bioinformatics 18, 1124–1132 (2002)
Horn, F., Lau, A., Cohen, F.: Automated extraction of mutation data from the literature: application of MuteXt to G protein-coupled receptors and nuclear hormone receptors. Bioinformatics 20, 557–568 (2004)
Mika, S., Rost, B.: Protein names precisely peeled off free text. Bioinformatics 20, I241–I247 (2004)
Mika, S., Rost, B.: NLProt: extracting protein names and sequences from papers. Nucleic Acids Res. 32, W634–W637 (2004)
Donaldson, I., Martin, J., de Bruijn, B., Wolting, C., Lay, V., Tuekam, B., Zhang, S., Baskin, B., Bader, G., Michalickova, K., Pawson, T., Hogue, C.: PreBIND and Textomy–mining the biomedical literature for protein-protein interactions using a support vector machine. BMC Bioinformatics 4, 11 (2003)
Bader, G., Betel, D., Hogue, C.: BIND: the Biomolecular Interaction Network Database. Nucleic Acids Res. 31, 248–250 (2003)
Xenarios, I., Salwinski, L., Duan, X., Higney, P., Kim, S., Eisenberg, D.: DIP, the Database of Interacting Proteins: a research tool for studying cellular networks of protein interactions. Nucleic Acids Res. 30, 303–305 (2002)
Chen, H., Sharp, B.: Content-rich biological network constructed by mining PubMed abstracts. BMC Bioinformatics 5, 147 (2004)
Author information
Authors and Affiliations
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2005 Springer-Verlag Berlin Heidelberg
About this paper
Cite this paper
Malik, R., Siebes, A. (2005). CONAN: An Integrative System for Biomedical Literature Mining. In: Bento, C., Cardoso, A., Dias, G. (eds) Progress in Artificial Intelligence. EPIA 2005. Lecture Notes in Computer Science(), vol 3808. Springer, Berlin, Heidelberg. https://doi.org/10.1007/11595014_25
Download citation
DOI: https://doi.org/10.1007/11595014_25
Publisher Name: Springer, Berlin, Heidelberg
Print ISBN: 978-3-540-30737-2
Online ISBN: 978-3-540-31646-6
eBook Packages: Computer ScienceComputer Science (R0)