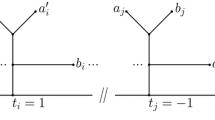
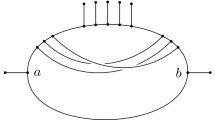
Abstract
We present a fast converging method for distance-based phylogenetic inference, which is novel in two respects. First, it is the only method (to our knowledge) to guarantee accuracy when knowledge about the model tree, i.e bounds on the edge lengths, is not assumed. Second, our algorithm guarantees that, with high probability, no false assertions are made. The algorithm produces a maximal forest of the model tree, in time Õ(n 3) in the typical case. Empirical testing has been promising, comparing favorably to Neighbor Joining, with the advantage of making few or no false assertions about the topology of the model tree; guarantees against false positives can be controlled as a parameter by the user.
Research supported by CIPRES (NSF ITR grant # NSF EF 03-31494).
Preview
Unable to display preview. Download preview PDF.
Similar content being viewed by others
References
Buneman, P.: The recovery of trees from measures of dissimilarity. In: Mathematics in the Archaeological and Historical Sciences, pp. 387–395. Edinburgh University Press, Edinburgh (1971)
Cavender, J.: Taxonomy with confidence. Mathematical Biosciences 40, 271–280 (1978)
Day, W.: Optimal algorithms for comparing trees with labelled leaves. J. Class. 2, 7–28 (1995)
Erdos, P., Steel, M., Szekely, L., Warnow, T.: A few logs suffice to build (almost) all trees (part 1). Random Structures and Algorithms 14(2), 153–184 (1999)
Erdos, P., Steel, M., Szekely, L., Warnow, T.: A few logs suffice to build (almost) all trees (part 2). Theoretical Computer Science 221, 77–118 (1999)
Farris, J.: A probability model for inferring evolutionary trees. Systematic Zoology 22, 250–256 (1973)
Golumbic, M.: Algorithmic Graph Theory and Perfect Graphs. Academic Press, New York (1980)
Huson, D., Nettles, S., Warnow, T.: Disk-Covering, A fast converging method for phylogenetic tree reconstruction. Journal of Computational Biology 6, 369–386 (1999)
Mossel, E.: Distorted metrics on trees and phylogenetic forests. IEEE Comp. Biol. and Bioinformatics (to appear, 2004), Availible at: http://arxiv.org/abs/math.CO/0403508
Mossel, E.: Phase Transitions in Phylogeny. Trans. Amer. Math. Soc. 356(6), 2379–2404 (2004) (electronic)
Neyman, J.: Molecular studies of evolution: a source of novel statistical problems. In: Gupta, S., Yackel, J. (eds.) Statistical Decision Theory and Related Topics. Academic Press, New York (1971)
Saitou, N., Nei, M.: The neighbor-joing method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425 (1987)
Usman, R., Moret, B., Warnow, T., Williams, T.: Rec-I-DCM3: A fast algorithmic technique for reconstructing large phylogenetic trees. In: Proc. IEEE Computer Society Bioinformatics Conference CSB 2004. Stanford Univ. (2004)
Author information
Authors and Affiliations
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2006 Springer-Verlag Berlin Heidelberg
About this paper
Cite this paper
Daskalakis, C., Hill, C., Jaffe, A., Mihaescu, R., Mossel, E., Rao, S. (2006). Maximal Accurate Forests from Distance Matrices. In: Apostolico, A., Guerra, C., Istrail, S., Pevzner, P.A., Waterman, M. (eds) Research in Computational Molecular Biology. RECOMB 2006. Lecture Notes in Computer Science(), vol 3909. Springer, Berlin, Heidelberg. https://doi.org/10.1007/11732990_24
Download citation
DOI: https://doi.org/10.1007/11732990_24
Publisher Name: Springer, Berlin, Heidelberg
Print ISBN: 978-3-540-33295-4
Online ISBN: 978-3-540-33296-1
eBook Packages: Computer ScienceComputer Science (R0)