Abstract
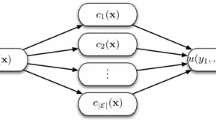
High dimensionality two-class biomarker data (e.g. microarray and proteomics data with few samples but large numbers of variables) is often difficult to classify. Many currently used methods cannot easily deal with unbalanced datasets (when the number of samples in class 1 and class 2 are very different). This problem can be alleviated by the following new method: first, sample data space by recursive partitions, then use two-class support vector machine tree (2-SVMT) for classification. Recursive partitioning divides the feature space into more manageable portions, from which informative features are more easily found by 2-SVMT. Using two-class microarray and proteomics data for cancer diagnostics, we demonstrate that 2-SVMT results in higher classification accuracy and especially more consistent classification of various datasets than standard SVM, KNN or C4.5. The advantage of the method is its super robustness for class unbalanced datasets.
Preview
Unable to display preview. Download preview PDF.
Similar content being viewed by others
References
Alon, U., Barkai, N., Notterman, D.A., Gish, K., Ybarra, S., Mack, D., Levine, A.J.: Broad Patterns of Gene Expression Revealed by Clustering Analysis of Tumour and Normal Colon Tissues Probed by Oligonucleotide Arrays. Proc Natl Acad Sci U. S. A. 96(12), 6745–6750 (1999)
Ein-Dor, L., Kela, I., Getz, G., Givol, D., Domany, E.: Outcome Signature Genes in Breast Cancer: Is There a Unique Set. Bioinformatics 21(2), 171–178 (2005)
Deb, K., Agrawal, S., Pratap, A., Meyarvian, T.: A Fast and Elitist Multi-objective Genetic Algorithm: NSGA-II. IEEE Transactions on Evolutionary Computation 6(2), 182–197 (2002)
Chawla, N.V., Bowyer, K.W., Hall, L.O., Kegelmeyer, W.P.: SMOTE, Synthetic Minority Over-sampling Technique. Journal of Artificial Intelligence Research 16, 321–357 (2002)
Kasabov, N.K.: Evolving Connectionist Systems, Methods and Applications in Bioinformatics, Brain Study and Intelligent Machines. Springer, Heidelberg (2002)
Pang, S., Kim, D., Bang, S.Y.: Face Membership Authentication Using SVM Classification Tree Generated by Membership-based LLE Data Partition. IEEE Trans. on Neural Networks 16(2), 436–446 (2005)
Ho, T.K.: The Random Subspace Method for Constructing Decision Forests. IEEE Transaction on Pattern Analysis and Machine Intelligence 20(8), 832–844 (1998)
Author information
Authors and Affiliations
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2006 Springer-Verlag Berlin Heidelberg
About this paper
Cite this paper
Pang, S., Havukkala, I., Kasabov, N. (2006). Two-Class SVM Trees (2-SVMT) for Biomarker Data Analysis. In: Wang, J., Yi, Z., Zurada, J.M., Lu, BL., Yin, H. (eds) Advances in Neural Networks - ISNN 2006. ISNN 2006. Lecture Notes in Computer Science, vol 3973. Springer, Berlin, Heidelberg. https://doi.org/10.1007/11760191_92
Download citation
DOI: https://doi.org/10.1007/11760191_92
Publisher Name: Springer, Berlin, Heidelberg
Print ISBN: 978-3-540-34482-7
Online ISBN: 978-3-540-34483-4
eBook Packages: Computer ScienceComputer Science (R0)