Abstract
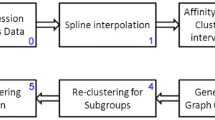
We focus on clustering gene expression temporal profiles, and propose a novel, simple algorithm that is powerful enough to find an efficient distribution of genes over clusters. We also introduce a variant of a clustering index that can effectively decide upon the optimal number of clusters for a given dataset. The clustering method is based on a profile-alignment approach, which minimizes the mean-square-error of the first order differentials, to hierarchically cluster microarray time-series data. The effectiveness of our algorithm has been tested on datasets drawn from standard experiments, showing that our approach can effectively cluster the datasets based on profile similarity.
Preview
Unable to display preview. Download preview PDF.
Similar content being viewed by others
References
Brazma, A., Vilo, J.: Gene expression data analysis. FEBS Lett. 480, 17–24 (2000)
Bréhélin, L.: Clustering Gene Expression Series with Prior Knowledge. In: Casadio, R., Myers, G. (eds.) WABI 2005. LNCS (LNBI), vol. 3692, pp. 27–38. Springer, Heidelberg (2005)
Chu, S., DeRisi, J., Eisen, M., Mulholland, J., Botstein, D., Brown, P., Herskowitz, I.: The transcriptional program of sporulation in budding yeast. Science 282, 699–705 (1998)
Drăghici, S.: Data Analysis Tools for DNA Microarrays. Chapman & Hall, Boca Raton (2003)
Duda, R., Hart, P., Stork, D.: Pattern Classification, 2nd edn. John Wiley and Sons, Inc., New York (2000)
Eisen, M., Spellman, P., Brown, P., Botstein, D.: Cluster analysis and display of genome-wide expression patterns. In: Proc. Natl Acad. Sci., USA, vol. 95, pp. 14863–14868 (1998)
Heyer, L., Kruglyak, S., Yooseph, S.: Exploring expression data: identification and analysis of coexpressed genes. Genome Res. 9, 1106–1115 (1999)
Iyer, V., Eisen, M., Ross, D., Schuler, G., Moore, T., Lee, J., Trent, J., Staudt Jr., L., Hudson, J., Boguski, M.: The transcriptional program in the response of human fibroblasts to serum. Science 283, 83–87 (1999)
Maulik, U., Bandyopadhyay, S.: Performance Evaluation of Some Clustering Algorithms and Validity Indices. IEEE Transactions on Pattern Analysis and Machine Intelligence 24(12), 1650–1654 (2002)
Peddada, S., Lobenhofer, E., Li, L., Afshari, C., Weinberg, C., Umbach, D.: Gene selection and clustering for time-course and dose-response microarray experiments using order-restricted inference. Bioinformatics 19(7), 834–841 (2003)
Rueda, L., Bari, A.: Clustering Microarray Time-Series Data Using a Mean-Square-Error Profile Alignment Algorithm (submitted for publication), Electronically available at: http://www.inf.udec.cl/~lrueda/papers/ProfileMSE-Jnl.pdf
Sherlock, G.: Analysis of large-scale gene expression data. Curr. Opin. Immunol. 12, 201–205 (2000)
Author information
Authors and Affiliations
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2006 Springer-Verlag Berlin Heidelberg
About this paper
Cite this paper
Bari, A., Rueda, L. (2006). A New Profile Alignment Method for Clustering Gene Expression Data. In: Lamontagne, L., Marchand, M. (eds) Advances in Artificial Intelligence. Canadian AI 2006. Lecture Notes in Computer Science(), vol 4013. Springer, Berlin, Heidelberg. https://doi.org/10.1007/11766247_8
Download citation
DOI: https://doi.org/10.1007/11766247_8
Publisher Name: Springer, Berlin, Heidelberg
Print ISBN: 978-3-540-34628-9
Online ISBN: 978-3-540-34630-2
eBook Packages: Computer ScienceComputer Science (R0)