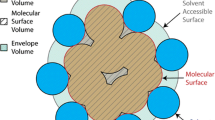
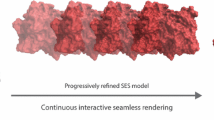
Abstract
The visualization of 3D volume data of proteins synthesized by quantum mechanics is a new topic and is of great importance in modern bio-computing. In this paper, we introduce our primary attempts on the volume visualization of the 3D macro-molecular scalar field. Firstly, we transform one protein molecular structure into a regularly sampled 3D scalar field according to the theories in quantum chemistry, in which each node records the combined effect of different actions in protease. We then exploit volume rendering techniques to find the macro-structure inside the data field based on a convenient mapping mechanism. We also propose an improved transfer function mode, facilitating the flexible visualization of the 3D protein data sets. Finally, combined with the iso-surface extraction technique, our approach allows for interactive exploration of the potential “tunnel” region which exhibits biological sense. With our approach, we show the escape route of water molecules hidden in the HIV-1 protease, which conforms to the experimental results.
Preview
Unable to display preview. Download preview PDF.
Similar content being viewed by others
References
Leach, A.R.: Molecular Modelling Principles and Applications. Prentice-Hall, Englewood Cliffs (2001)
Bajaj, C., Pascucci, V., Schikore, D.: The Contour Spectrum. In: Proceedings of the 1997 IEEE Visualization Conference, pp. 167–173.
Johnson, C.: Top Scientific Visualization Research Problems. IEEE Computer Graphics and Applications 24(4), 13–17 (2004)
Stan, C., Tsai: An introduction to computational biochemistry. Wiley-Liss, Inc., New York (2002)
Hoenigmann, D., Ruisz, J., Haider, C.: Adaptive Design of a Global Opacity Transfer Function for Direct Volume Rendering of Ultrasound Data. In: IEEE Visualization 2003, Seattle, Washington, USA, pp. 489–496 (2003)
Kindlmann, G., Whitaker, R., Tasdizen, T., Moeller, T.: Curvature-Based Transfer Functions for Direct Volume Rendering: Methods and Applications. In: IEEE Visualization 2003, Seattle, Washington, USA, pp. 513–520 (2003)
Kindlmann, G., Durkin, J.W.: Semi-Automatic Generation of Transfer Functions for Direct Volume Rendering. In: Proceedings of the 1998 Symposium on Volume visualization, pp. 79–86 (1998)
Gerig, G., Kindlemann, G., et al.: Image Processing for Volume Graphics. In: A Course for SIGGRAPH 2002 (2002)
Berman, H.M., Westbrook, J., Feng, Z., Gilliland, G., Bhat, T.N., Weissig, H., Shindya-lov, I.N., Bourne, P.E.: The Protein Data Bank. Nucleic Acids Research 28, 235–242 (2000)
Pfister, H., Lorensen, B., Bajaj, C., Kindlmann, G.: The Transfer Function Bake-Off. IEEE Computer Graphics and Applications 21(3), 16–22 (2001)
Levine, I.N.: Quantum Chemistry. Prentice-Hall, Englewood Cliffs (2004)
Kniss, J., Kindlmann, G., Hansen, C.: Multidimensional Transfer Functions for Interactive Volume Rendering. IEEE Transactions on Visualization and Computer Graphics 8(3), 270–285 (2002)
Kniss, J., Premǒze, S., Ikits, M., Lefohn, A., et al.: Gaussian Transfer Functions for Multi-Field Volume Visualization. In: IEEE Visualization 2003, Seattle, Washington, USA, pp. 497–504 (2003)
Engel, K., Kraus, M., Ertl, T.: High-Quality Pre-Integrated Volume Rendering Using Hard-ware-Accelerated Pixel Shading. In: Eurographics/SIGGRAPH Workshop on Graphics Hardware (2001)
Kniss, J.M., Van Uitert, R., Stephens, A., Li, G.-S., Tasdizen, T.: Statistically Quantitative Volume Visualization. In: IEEE Visualization 2005, Minneapolis, USA, pp. 287–294 (2005)
Levoy, M.: Display of Surfaces from Volume Data. IEEE CG & A 8(5), 29–37 (1988)
Nicoletti, G.M.: Volume visualization: advances in transfer and opacity function generation for interactive direct volume rendering. In: Proceedings of the Thirty-Sixth Southeastern Symposium on System Theory, pp. 1–5 (2004)
Mehta, S., Hazzard, K., Machiraju, R.: Detection and Visualization of Anomalous Structures in Molecular Dynamics Simulation Data. In: IEEE Visualization 2004, Texas, USA, pp. 465–472 (2004)
Takahashi, S., Takeshima, Y., Fujishiro, I.: Topological volume skeletonization and its application to transfer function design. Graphical Models 66, 24–49 (2004)
Potts, S., Möller, T.: Transfer Functions on a Logarithmic Scale for Volume Rendering. In: Proceedings of Graphics Interfaces 2004, London, Ontario, pp. 57–63 (May 2004)
Elvins, T.T.: A Survey of Algorithms for Volume Visualization. Computer Graphics 26(3), 194–201 (1992)
Qiao, W., Ebert, D.S., Entezari, A., Korkusinski, M., Klimeck, G.: VolQD: Direct Volume Rendering of Multi-million Atom Quantum Dot Simulations. In: IEEE Visualization 2005, Inneapolis, MN, USA, pp. 319–326 (October 2005)
Humphrey, W., Dalke, A., Schulten, K.: VMD - Visual Molecular Dynamics. Journal of Molecular Graphics 14(1), 33–38 (1996)
Tao, W.: Personal Communication
http://molvis.sdsc.edu/protexpl/frntdoor.htm (Last visiting data: 2006-1-12)
http://www.rasmol.org/ (Last visiting data: 2006-1-12)
Author information
Authors and Affiliations
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2006 Springer-Verlag Berlin Heidelberg
About this paper
Cite this paper
Hu, M., Chen, W., Zhang, T., Peng, Q. (2006). Direct Volume Rendering of Volumetric Protein Data. In: Nishita, T., Peng, Q., Seidel, HP. (eds) Advances in Computer Graphics. CGI 2006. Lecture Notes in Computer Science, vol 4035. Springer, Berlin, Heidelberg. https://doi.org/10.1007/11784203_34
Download citation
DOI: https://doi.org/10.1007/11784203_34
Publisher Name: Springer, Berlin, Heidelberg
Print ISBN: 978-3-540-35638-7
Online ISBN: 978-3-540-35639-4
eBook Packages: Computer ScienceComputer Science (R0)