Abstract
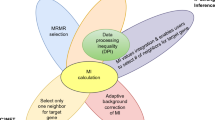
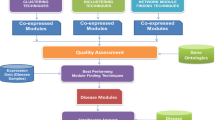
Finding gene relations has become important in research, since finding relations could assist biologists in finding a genes functionality. This article describes our proposal to combine microarray data and literature to find the relations among genes. The proposed method tries emphasizes the combined use of microarray data and literature rather than microarray data alone. Currently, many scholars use clustering algorithms to analyze microarray data, but these algorithms can find only the same expression mode, not the transcriptional relation between genes. Moreover, most traditional approaches involve all-against-all comparisons that are time-consuming. To reduce the comparison time and to find more relations in a microarray, we propose a method to expand microarray data and use association-rule algorithms to find all possible rules first. With its literature text mining, our method can be used to select the most suitable rules. Under such circumstances, the suitable gene group is selected and the gene comparison frequency is reduced sharply. Finally, we can then apply dynamic Bayesian network (DBN) to find the genes interaction. Unlike other techniques, this method not only reduces the comparison complexity but also reveals more mutual interactions among genes.
Preview
Unable to display preview. Download preview PDF.
Similar content being viewed by others
References
Becquet, C., Blachon, S., Jeudy, B., Boulicaut, J.F., Gandrillon, O.: Strong-association-rule mining for large-scale gene-expression data analysis: a case study o human SAGE data. Genome Biol. 12, 1–16 (2003)
Creighton, C., Hanash, S.: Mining gene expression databases for association rules. Bioinformatics 19, 79–86 (2003)
Doddi, S., Marathe, A., Ravi, S.S., Torney, D.C.: Discovery of association rules in medical data. Med. Inform. Internet Med. 26, 25–33 (2001)
Eisen, M.B., Spellman, P.T., Brown, P.O., Botstein, D.: Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Science (USA) 95, 14863–14868 (1998)
Eisenberg, D., Marcotte, M.E., Xenarios, I., Yeates, O.T.: Protein function in the post-genomic era. Nature 405, 823–826 (2000)
Ewing, B., Green, P.: Analysis of expressed sequence tags indicates 35,000 human genes. Nature Genet. 25, 232–234 (2000)
Hieter, P., Boguski, M.: Functional genomics: its all how you read it. Science 278, 601–602 (1997)
Jenssen, T., Lagreid, A., Komorowski, J., Hovig, E.: A literature network of human genes for high-throughput analysis of gene expression. Nature genetics 28, 21–28 (2001)
Ji, L., Tan, K.L.: Mining Gene expression data for positive and negative co-regulated gene cluster. Bioinformatics 20(16), 2711–2718 (2004)
Kim, S.Y., Imoto, S., Miyano, S.: Inferring gene networks from time series microarray data using Dynamic Bayesian Networks. Briefing Bioinformatics 4(3), 228–235 (2003)
Kim, S.Y., Imoto, S., Miyano, S.: Dynamic Bayesian networks and nonparametric regression for nonlinear modeling of gene networks from time series gene expression data. Biosystems 75, 57–65 (2004)
Murphy, K., Mian, S.: Modeling gene expression data using dynamic Bayesian networks. Technical Report, Computer Science Division. University of California, Berkeley, CA (1999)
Narayanasamy, V., Mukhopadhyay, S., Palakal, M., Potter, D.A.: TransMiner:Mining Transitive Associations among Biological Objects form Text. Journal of biomedical science 11, 864–873 (2004)
Ong, I.M., Glasner, J.D., Page, D.: Modeling regulatory pathways in E.coli from time series expression profiles. Bioinformatics 18, 241–248 (2002)
Salton, G., Wong, A., Yang Cornel, C.S.: A Vector Space Model for Automated Indexing. Journal of the ACM 18(1), 613–620 (1975)
Shatkay, H., Edwards, S., Boguski, M.: Information retrieval meets gene analysis. IEEE Intelligent Systems, Special Issue on Intelligent Systems in Biology 17(2), 45–53 (2002)
Stephens, M., Palakal, M., Mukhopadhyay, S., Raje, R., Mostafa, J.: Detecting gene relations from medline abstracts. Pac. Symp. Biocomput., 483–495 (2001)
Tamayo, P., Slonim, D., Mesirov, J., Zhu, Q., Kitareewan, S., Dmitrovsky, E., Lander, E.S., Golub, T.R.: Interpreting patterns of gene expression with self-organizing maps methods and application to hematopoietic differentiation. Nature Genetics 96, 2907–2912 (1999)
Tao, Y.C., Leibel, R.L.: Identifying functional relationships among human genes by systematic analysis of biological literature. BMC Bioinformatics 3(16), 1–9 (2002)
Tavazoie, S., Hughes, J.D., Campbell, M.J., Cho, R.J., Church, G.M.: Systematic determination of genetic network architecture. Nature Genetics 22, 281–285 (1999)
Torgeir, R.H., Astrid, L., Jan, K.: Learning rule-based models of biological process from gene expression time profiles using Gene Ontology. Bioinformatics 19, 1116–1123 (2002)
Webb, G.I., Zhang, S.: K-Optimal Rule Discovery. Data mining and Knowledge Discovery 10(1), 39–79 (2005)
Zou, M., Conzen, S.D.: A new dynamic Bayesian network approach for identifying gene regulatory networks from time course microarray data. Bioinformatics, Advance Access published on August 12, 2004, pp. 1–29 (2004)
Author information
Authors and Affiliations
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2006 Springer-Verlag Berlin Heidelberg
About this paper
Cite this paper
Wang, HC., Lee, YS., Huang, TH. (2006). Gene Relation Finding Through Mining Microarray Data and Literature. In: Priami, C., Hu, X., Pan, Y., Lin, T.Y. (eds) Transactions on Computational Systems Biology V. Lecture Notes in Computer Science(), vol 4070. Springer, Berlin, Heidelberg. https://doi.org/10.1007/11790105_7
Download citation
DOI: https://doi.org/10.1007/11790105_7
Publisher Name: Springer, Berlin, Heidelberg
Print ISBN: 978-3-540-36048-3
Online ISBN: 978-3-540-36049-0
eBook Packages: Computer ScienceComputer Science (R0)