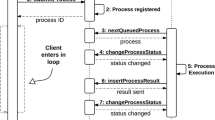
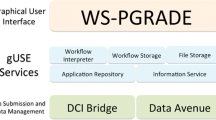
Abstract
Our workflow platform offers a view of the different tools available as a single and uniform pool of services readily available for enhancing query processing. This proposal is based on an architecture for publishing biological data and services, and is designed to be a flexible client for making use of BioMOBY servers, extending them with persistency of the information retrieved for each user. We also present in this paper some biological results, which have been obtained by taking advantage of the proposed workflow execution system. This work has been developed and implemented in the National Institute for Bioinformatics (INB) in Spain (available at http://www.inab.org/MOWServ).
Preview
Unable to display preview. Download preview PDF.
Similar content being viewed by others
References
Stevens, R.D., Baker, P.G., Bechhofer, S., Ng, G., Jacoby, A., Paton, N., Goble, C.A., Brass, A.: TAMBIS: Transparent Access to Multiple Bioinformatics Information Sources. Bioinformatics 16(2), 184–186 (2000)
Ludäscher, B., Gupta, A., Martone, M.E.: A Model Based Mediator System for Scientific Data Management. In: Lacroix, Z., Critchlow, T. (eds.) Bioinformatics: Managing Scientific Data, pp. 335–370 (2003)
Lange, M., Freier, A., Scholz, U., Stephanik, A.: A computational Support for Access to Integrated Molecular Biology Data (2001)
Gracy, J., Chiche, L.: PAT: a protein analysis toolkit for integrated biocomputing on the web. Nucl. Acids Res. 33, W65–W71 (2005)
Aldana, J.F., Hidalgo-Conde, M., Navas, I., Roldán, M.M., Trelles, O.: Bio-Broker: A biological data and services mediator system. In: IADIS International Conference, Applied Computing 2005, Algarve, Portugal, February 22-25, pp. 527–534 (2005)
Wilkinson, M.D., Gessler, D., Farmer, A., Stein, L.: The Bio-MOBY Project Explores Open-Source, Simple, Extensible Protocols for Enabling Biological Database Interoperability. In: Proceedings Virtual Conference Genomic and Bioinformatics, vol. (3), pp. 16–26 (2003) ISSN 1547-383X
Oinn, T., Addis, M., Ferris, J., Marvin, D., Senger, M., Greenwood, M., Carver, T., Glover, K., Pocock, M.R., Wipat, A., Li, P.: Taverna: A tool for the composition and enactment of bioinformatics workflows. Bioinformatics Journal 20(17), 3045–3054 (2004)
Ponting, C.P.: Tudor domains in proteins that interact with RNA. Trends Biochem. Sci. 22, 51–52 (1997)
Author information
Authors and Affiliations
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2006 Springer-Verlag Berlin Heidelberg
About this paper
Cite this paper
Navas-Delgado, I., Pérez, A.J., Aldana-Montes, J.F., Trelles, O. (2006). Distributed Execution of Workflows in the INB. In: Leser, U., Naumann, F., Eckman, B. (eds) Data Integration in the Life Sciences. DILS 2006. Lecture Notes in Computer Science(), vol 4075. Springer, Berlin, Heidelberg. https://doi.org/10.1007/11799511_20
Download citation
DOI: https://doi.org/10.1007/11799511_20
Publisher Name: Springer, Berlin, Heidelberg
Print ISBN: 978-3-540-36593-8
Online ISBN: 978-3-540-36595-2
eBook Packages: Computer ScienceComputer Science (R0)