Abstract
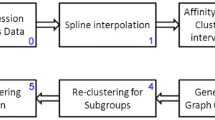
Microarray experiments produce large data sets that often contain noise and considerable missing data. Typical clustering methods such as hierarchical clustering or partitional algorithms can often be adversely affected by such data. This paper introduces a method to overcome such problems associated with noise and missing data by modelling the time series data with polynomials and using these models to cluster the data. Similarity measures for polynomials are given that comply with commonly used standard measures. The polynomial model based clustering is compared with standard clustering methods under different conditions and applied to a real gene expression data set. It shows significantly better results as noise and missing data are increased.
Preview
Unable to display preview. Download preview PDF.
Similar content being viewed by others
References
Altman, D.G.: Practical Statistics for Medical Research. Chapman and Hall, Boca Raton (1997)
Bozdech, Z., Llinás, M., Pulliam, B.L., Wong, E.D., Zhu, J., DeRisi, J.L.: The Transcriptome of the Intraerythrocytic Developmental Cycle of Plasmodium falciparum. PLoS Biology 1, 85–100 (2003)
Eisen, M.B., Spellman, P.T., Brown, P.O., Botstein, D.: Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA 95, 14863–14868 (1998)
Hand, D.J., Mannila, H., Smyth, P.: Principles of Data Mining. MIT Press, Cambridge (2001)
Jain, A.K., Murty, M.N., Flynn, P.J.: Data Clustering: A Review. ACM Computer Surveys 32(3), 264–323 (1999)
Kaufman, L., Rousseeuw, P.J.: Clustering by means of Medoids. In: Dodge, Y. (ed.) Statistical Data Analysis based on the L1-Norm, pp. 405–416. North-Holland, Amsterdam (1987)
Kellam, P., Liu, X., Martin, N., Orengo, C., Swift, S., Tucker, A.: Comparing, Contrasting and Combining Clusters in Viral Gene Expression Data. In: Proceedings of the IDAMAP 2001 Workshop, London, pp. 56–62 (2001)
Lichtenberg, G., Faisal, S., Werner, H.: Ein Ansatz zur dynamischen Modellierung der Genexpression mit Shegalkin-Polynomen (An Approach to Dynamic Modelling of Gene Expression by Zhegalkin Polynomials). at – Automatisierungstechnik 53(12), 589–596 (2005)
Ralston, A.: A First Course in Numerical Analysis. McGraw-Hill, New York (1965)
Spellman, P.T., Sherlock, G., Zhang, M.Q., Iyer, V.R., Anders, K., Eisen, M.B., Brown, P.O., Botstein, D., Futcher, B.: Comprehensive Identification of Cell Cycle-regulated Genes of the Yeast Saccharomyces cerevisiae by Microarray Hybridization. Molecular Biology of the Cell 9, 3273–3297, URL: http://cellcycle-www.stanford.edu
Stekel, D.: Microarray Bioinformatics. Cambridge University Press, Cambridge (2003)
Vinciotti, V., Liu, X., Turk, R., de Meijer, E.J., t’ Hoen, P.A.C.: Exploiting the full power of temporal gene expression profiling through a new statistical test: Application to the analysis of muscular dystrophy data. BMC Bioinformatics 7, 183 (2006)
Wit, E., McClure, J.: Statistics for Microarrays. John Wiley, Chichester (2004)
Author information
Authors and Affiliations
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2006 Springer-Verlag Berlin Heidelberg
About this paper
Cite this paper
Hirsch, M. et al. (2006). Improved Robustness in Time Series Analysis of Gene Expression Data by Polynomial Model Based Clustering. In: R. Berthold, M., Glen, R.C., Fischer, I. (eds) Computational Life Sciences II. CompLife 2006. Lecture Notes in Computer Science(), vol 4216. Springer, Berlin, Heidelberg. https://doi.org/10.1007/11875741_1
Download citation
DOI: https://doi.org/10.1007/11875741_1
Publisher Name: Springer, Berlin, Heidelberg
Print ISBN: 978-3-540-45767-1
Online ISBN: 978-3-540-45768-8
eBook Packages: Computer ScienceComputer Science (R0)