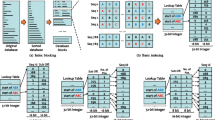
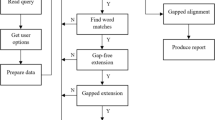
Abstract
BLAST is a tool for finding biologically similar sequences to given query sequences in annotated sequence database. Since the number of sequences in the database increases at exponential rate, and the number of users drastically increases, the performance of BLAST is a primary concern to service sites like NCBI. NCBI developed a parallel BLAST for the speedup of BLAST using threads on SMP machines. But the speedup is still limited due to the architectural limitations of SMP machines. Distributed memory multiprocessor can be an alternative choice for cost-effective search in very large scale sequence data. However for an optimized configuration of Cluster systems and SMP machines, the performance study of BLAST on SMP machines is essential. In this paper, we analyze BLAST and BLAST algorithms to enhance the performance of BLAST on parallel machines and report the performance of BLAST on SMP machines. Some important runtime characteristics of BLAST are identified through the performance evaluation. According to our performance test, PC clusters or clusters of low-way SMP machines outperform high-way SMP machines in terms of cost-effectiveness. Besides, BLAST on Linux operating system shows better performance than BLAST on Solaris operating system in the same configurations.
Preview
Unable to display preview. Download preview PDF.
Similar content being viewed by others
References
NCBI: Growth of GenBank. Technical report, National Center for Biotechnology Information (March 12, 2002)
Chi, E.H., Shoop, E., Carlis, J., Retzel, E., Ried, J.: Efficiency of shared-memory multiporceossors for a genetic sequence similiarity search algorithm. Technical report, Computer Science Dept., University of Minnesota (1997)
Altschul, S.F., Gish, W., Miller, W., Myers, E.W., Lipman, D.J.: Basic Local Alignment Search Tool. Journal of Molecular Biology 215, 403–410 (1990)
Altschul, S., Gish, W.: Local alignment statistics. Methods in Enzymology 266, 460–480 (1996)
Altschul, S., Madden, T., Schaffer, A., Zhang, J., Zhang, Z., Miller, W., Lipman, D.: Gapped BLAST and PSI–BLAST: A new generation of protein katabase search programs. Nucleic Acids Research 25, 3389–3402 (1997)
Camp, N., Cofer, H., Gomperts, R.: High-Throughput BLAST. Technical report, Silicon Graphics, Inc. (1998)
Hwang, K., Xu, Z.: Chapter 12 Parallel Paradignms and Programming Model. In: Scalable Parallel Computing, McGraw-Hill Companies, Inc., New York (1998)
Author information
Authors and Affiliations
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2006 Springer-Verlag Berlin Heidelberg
About this paper
Cite this paper
Kim, HS., Kim, HJ., Han, DS. (2006). Performance Evaluation of BLAST on SMP Machines. In: Min, G., Di Martino, B., Yang, L.T., Guo, M., Rünger, G. (eds) Frontiers of High Performance Computing and Networking – ISPA 2006 Workshops. ISPA 2006. Lecture Notes in Computer Science, vol 4331. Springer, Berlin, Heidelberg. https://doi.org/10.1007/11942634_69
Download citation
DOI: https://doi.org/10.1007/11942634_69
Publisher Name: Springer, Berlin, Heidelberg
Print ISBN: 978-3-540-49860-5
Online ISBN: 978-3-540-49862-9
eBook Packages: Computer ScienceComputer Science (R0)