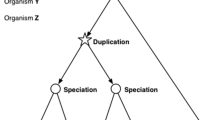
Abstract
Bioinformatics databases are heterogeneous, differ in their representation as well as in their query capabilities across diverse information held in distributed autonomous resources. Current approaches to integrating heterogeneous bioinformatics data sources are based on one of a: common field, ontology or cross-reference. In this paper we investigate the use of semantic relationships across species to link, integrate and annotate genes from publicly available data sources and a novel Soft Link approach is introduced, to link information across species held in biological databases, through providing a flexible method of joining related information from different databases, including non-bioinformatics databases. A measure of relationship closeness will afford a biologist a new tool in their repertoire for analysis. Soft Links are identified as interrelated concepts and can be used to create a rich set of possible relation types supporting the investigation of alternative hypothesis.
Preview
Unable to display preview. Download preview PDF.
Similar content being viewed by others
References
Aparicio, A.S., Farias, O.L.M., et al.: Applying Ontologies in the Integration of Heterogeneous Relational Databases. In: Australasian Ontology Workshop (AOW 2005), Sydney, Australia, ACS (2005)
Baxevanis, A.D., Ouellette, B.F.F. (eds.): Bioinformatics: A Practical Guide to the Analysis of Genes and Proteins. John Wiley & Sons, New York (2001)
Ben-Miled, Z., Li, N., et al.: On the Integration of a Large Number of Life Science Web Databases. Lecture Notes in Bioinformatics (LNBI), pp. 172–186 (2004)
Ben Milad, Z., Liu, Y., et al.: Distributed Databases (2003)
Bleiholder, J., Lacroix, Z.e., et al.: BioFast: Challenges in Exploring Linked Life Science Sources. SIGMOD Record 33(2), 72–77 (2004)
Carel, R.: Practical Data Integration In Biopharmaceutical Research and Development. PharmaGenomics, 22–35 (2003)
Collet, C., Huhns, M.N., et al.: Resource Integration Using a Large Knowledge Base in Carnot. IEEE Computer 24(12), 55–62 (1991)
Davidson, S., Crabtree, J., et al.: K2/Kleisli and GUS: experiments in integrated access to genomic data sources. IBM Journal (2001)
Decker, S., Erdmann, M., et al.: Ontobroker: Ontology Based Access to Distributed and Semi-Structured Information. Database Semantics - Semantic Issues in Multimedia Systems. In: Proceedings TC2/WG 2.6 8th Working Conference on Database Semantics (DS-8), Rotorua, New Zealand. Kluwer Academic Publishers, Boston (1999)
Dennis Jr., G., Sherman, B.T., et al.: DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 4(5), P3 (2003)
Etzold, T., Ulyanov, A., et al.: SRS: information retrieval system for molecular biology data banks. Methods Enzymol. 266, 114–128 (1996)
Freier, A., Hofestadt, R., et al.: BioDataServer: a SQL-based service for the online integration of life science data. Silico Biol. 2(2), 37–57 (2002)
Goble, C., Stevens, R., et al.: Transparent Access to Multiple Bioinformatics Information Sources. IBM Systems Journal 40(2), 534–551 (2001)
Gruber, T.R.: Toward principles for the design of ontologies used for knowledge sharing. International Journal of HumanComputer Studies 43, 907–928 (1995)
Gupta, A., Ludäscher, B., et al.: Knowledge-Based Integration of Neuroscience Data Sources. In: 12th International Conference on Scientific and Statistical Database Management (SSDBM), Berlin, Germany. IEEE Computer Society Press, Los Alamitos (2000)
Heflin, J., Hendler, J.: Dynamic Ontologies on the Web. In: Proceedings of 17th National Conference on Artificial Intelligence (AAAI 2000), Menlo Park,CA. AAAI/MIT Press (2000)
Kashyap, V., Sheth, A.P.: Semantic and schematic similarities between database objects: A context-based approach. VLDB Journal: Very Large Data Bases 5(4), 276–304 (1996)
Lacroix, Z., Critchlow, T. (eds.): Bioinformatics: Managing Scientific Data. Multimedia information and systems. Morgan Kaufmann, San Francisco (2003)
Leser, U., Naumann, F.: (Almost) Hands-Off Information Integration for the Life Sciences. In: Proceedings of the Conference in Innovative Database Research (CIDR) 2005, Asilomar, CA (2005)
Necib, C.B., Freytag, J.C.: Using Ontologies for Database Query Reformulation. In: ADBIS (Local Proceedings) (2004)
Rector, A., Bechhofer, S., et al.: The grail concept modelling language for medical terminology. Artificial Intelligence in Medicine 9, 139–171 (1997)
Robert, H., Patricia, M.: SRS as a possible infrastructure for GBIF. GBIF DADI Meeting, San Diego (2002)
Venkatesh, T.V., Harlow, H.: Integromics: challenges in data integration. Genome Biology 3(8), reports4027.1 – reports4027.3 (2002)
Wache, H., Ogele, T.V., et al.: Ontology-Based Integration of Information — A Survey of Existing Approaches. In: IJCAI 2001 Workshop on Ontologies and Information Sharing, Seattle, USA. (2001)
Wiederhold, G.: Mediators in the architecture of future information systems. Computer 25(3), 38–49 (1992); The Genomics Unified Schema(GUS) platform for Functional genomics (2004)
Al-Daihani, B., Gray, A., et al.: Soft Link Model(SLM) for Bioinformatics Data Source Integration. In: International Symposium on Health Informatics and Bioinformatics, Turkey 2005, Antalya, Turkey, Middle East Technical University (2005)
Ashburner, M., Ball, C.A., et al.: Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 25(1), 25–29 (2000)
Benson, D.A., Karsch-Mizrachi, I., et al.: GenBank. Nucleic Acids Res 33(Database issue), D34–D38 (2005)
Bleiholder, J., Lacroix, Z.e., et al.: BioFast: Challenges in Exploring Linked Life Science Sources. SIGMOD Record 33(2), 72–77 (2004)
Buntrock, R.E.: Chemical registries–in the fourth decade of service. J. Chem. Inf. Comput. Sci. 41(2), 259–263 (2001)
Etzold, T., Ulyanov, A., et al.: SRS: information retrieval system for molecular biology data banks. Methods Enzymol. 266, 114–128 (1996)
Freier, A., Hofestadt, R., et al.: BioDataServer: a SQL-based service for the online integration of life science data. Silico Biol. 2(2), 37–57 (2002)
Gupta, A., Ludäscher, B., et al.: Knowledge-Based Integration of Neuroscience Data Sources. In: 12th International Conference on Scientific and Statistical Database Management (SSDBM), Berlin, Germany. IEEE Computer Society Press, Los Alamitos (2000)
Kanz, C., Aldebert, P., et al.: The EMBL Nucleotide Sequence Database. Nucleic Acids Res. 33(Database issue), D29–D33 (2005)
Kohler, J.: SEMEDA: Ontology based semantic integration of biological databases (2003)
Kohler, J.: Integration of life science databases. BioSlico 2(2), 61–69 (2004)
Lacroix, Z., Critchlow, T. (eds.): Bioinformatics: Managing Scientific Data. Multimedia information and systems. Morgan Kaufmann, San Francisco (2003)
Leser, U., Naumann, F.: (Almost) Hands-Off Information Integration for the Life Sciences. In: Proceedings of the Conference in Innovative Database Research (CIDR) 2005, Asilomar, CA (2005)
Letovsky, S.L. (ed.): Bioinformatics: databases and systems. Kluwer Academic Publishers, Massachusetts (1999)
Maglott, D., Ostell, J., et al.: Entrez Gene: gene-centered information at NCBI. Nucleic Acids Res. 33(Database issue), D54–D58 (2005)
Robbins, R.J.: Information infrastructure for the human genome project. IEEE Engineering in Medicine and Biology 14(6), 746–759 (1995)
Schneider, M., Tognolli, M., et al.: The Swiss-Prot protein knowledgebase and ExPASy: providing the plant community with high quality proteomic data and tools. Plant Physiol Biochem. 42(12), 1013–1021 (2004)
Williams, N.: How to get databases talking the same language. Science 275(5298), 301–302 (1997)
Barrett, T., Suzek, T.O., et al.: NCBI GEO: mining millions of expression profiles–database and tools. Nucl. Acids Res. %R 10.1093/nar/gki022 33(suppl. 1), D562–D566 (2005)
Lord, P.W., Stevens, R.D., et al.: Investigating semantic similarity measures across the Gene Ontology: the relationship between sequence and annotation. Bioinformatics %R 10.1093/bioinformatics/btg153 19(10), 1275–1283 (2003)
Author information
Authors and Affiliations
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2006 Springer-Verlag Berlin Heidelberg
About this paper
Cite this paper
Al-Daihani, B., Gray, A., Kille, P. (2006). Bioinformatics Data Source Integration Based on Semantic Relationships Across Species. In: Dalkilic, M.M., Kim, S., Yang, J. (eds) Data Mining and Bioinformatics. VDMB 2006. Lecture Notes in Computer Science(), vol 4316. Springer, Berlin, Heidelberg. https://doi.org/10.1007/11960669_8
Download citation
DOI: https://doi.org/10.1007/11960669_8
Publisher Name: Springer, Berlin, Heidelberg
Print ISBN: 978-3-540-68970-6
Online ISBN: 978-3-540-68971-3
eBook Packages: Computer ScienceComputer Science (R0)